梅花鹿(Cervus nippon)属于鹿科鹿属的中型反刍动物,其产品(如鹿茸)具有较高的药用价值。与其他反刍动物一样,梅花鹿依赖瘤胃微生物将结构性碳水化合物降解为挥发性脂肪酸为机体提供能量。在自然条件下,梅花鹿采食范围广,而且对一些含有毒素的植物(如狼毒花、飞燕草和富含单宁枝叶等)具有耐受性[1],这表明梅花鹿瘤胃微生物可能具有一定独特性。杨镒峰等[2]研究表明,饲粮中添加2%单宁可提高梅花鹿蛋白质代谢率。Hiura等[3]发现北海道地区野生梅花鹿也喜采食单宁含量高的树叶,并从瘤胃中分离到可有效降解单宁的链球菌属(Streptococcus spp.)。Pope等[4]和Sundset等[5, 6]对挪威地区驯鹿瘤胃微生物区系研究表明,驯鹿瘤胃内细菌主要由拟杆菌门和厚壁菌门细菌组成,细菌区系与其他反刍动物不同。而且Shi等[7]证明动物种类对瘤胃微生物有显著影响。牦牛、大额牛、晋南牛和羊驼的瘤胃细菌多样性的研究已有报道[8, 9, 10, 11],但基于非培养技术分析梅花鹿瘤胃细菌区系的研究却较少。本研究对以柞树叶为主要粗饲料的梅花鹿瘤胃细菌16S rRNA基因序列进行分析,研究梅花鹿瘤胃细菌区系,为丰富瘤胃微生物资源和揭示梅花鹿耐受单宁机制提供基础依据。
选用2头2岁龄的装有永久性瘤胃瘘管的成年雄性梅花鹿,平均体重130 kg,饲养于中国农业科学院特产研究所茸鹿实验基地(北纬44.04°,东经129.09°)。每天09:00和16:00定时定量各饲喂1次,以放牧状态下梅花鹿喜欢采食的柞树叶为主要粗饲料(柞树叶单宁含量为0.973%),饲粮组成及营养水平见表1。持续饲喂30 d(2011年10月1日至2011年10月31日)。第31天09:00饲喂之前通过瘤胃瘘管取瘤胃内容物,置于冰盒中带回实验室,保存于-80 °C冰箱备用。
| 表1 饲粮组成及营养水平(风干基础) Table 1 Composition and nutrient levels of the diet (air-dry basis) % |
PCR相关试剂和酶购于TaKaRa公司;PCR引物由上海生工生物工程有限公司合成;琼脂糖凝胶纯化回收试剂盒(QIAEX Ⅱ Gel Extraction Kit)购于QIAGEN公司;克隆转化试剂盒(TOPO TA Cloning Kit)购于Invitrogen公司。
细菌基因组DNA提取按照Lamontagne等[12]和郑刚等[13]方法进行,略作修改。取0.5 g瘤胃固液混合物,加入800 μL溶菌酶溶液 和20 μL蛋白酶K(20 mg/mL)。37 ℃、220 r/min震荡1 h后,加入300 μL裂解液 、300 μL磷酸盐缓冲液(pH 8.0)和600 μL苯酚∶ 氯仿∶ 异戊醇(25∶ 24∶ 1)溶液,65 ℃水浴30 min,每隔10 min在涡旋振荡仪上振荡30 s。4 ℃、5 000 r/min离心6 min,上清中加入0.6倍体积异丙醇,-80 ℃沉淀15 min。70%乙醇洗涤沉淀2~3次。适量TE缓冲液(pH 8.0)溶解粗提DNA,试剂盒纯化粗提基因组DNA。
细菌通用引物27F(5′-AGAGTTTGATCMTGGCTCAG-3′)和1492R(5′-TACGGYTACCTTGTTACGACTT-3′)扩增细菌16S rRNA基因[14]。反应体系50 μL,反应条件如下:95 ℃ 5 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 2 min(30个循环);72 ℃ 10 min。利用琼脂糖凝胶纯化回收试剂盒纯化PCR产物。将来自2头动物的等体积(10 μL)PCR产物混合,按照试剂盒(TOPO TA Cloning Kit)说明书构建16S rRNA基因克隆文库。随机挑选阳性克隆,用引物M13-47(5′-CGCCAGGGTTTTCCCAGTCACGAC-3′)和RV-M(5′-GAGCGGATAACAATTTCACACAGG-3′)对阳性克隆进行验证,确认PCR片段连接到载体。用上游引物27F对阳性克隆进行单向测序,测序由上海生工生物工程有限公司完成。
Bellerophon软件去除可能嵌合体序列[15]。使用Ribosomal Database Project (RDP) Release 10(RDP10)中Classifer程序对所有序列分类[16]。MOTHUR软件划分分类操作单元(OTU)[17],将相似性大于97%的序列视为同一个OTU[18]。选取每个OTU代表序列,利用BLAST程序在NCBI中搜索相似性最高序列[19]。以嗜火产液菌(Aquifex pyrophilus)为外群,利用MEGA 5.05软件中ClustalW比对后输出为同一长度序列,利用Kimura-two参数矩阵模型和邻接法(neighbor-joining,NJ)进行系统发育分析,设置Bootstrap值为1 000[20]。
本试验共获得128个16S rRNA基因序列,其中21个为可能嵌合体序列。RDP分类表明107个克隆序列代表拟杆菌门和厚壁菌门细菌,所占比例分别为87.9%和12.1%。以97%序列相似性为阈值,107个序列分为22个OTUs(表2)。MOTHUR计算表明克隆文库库容值为84.3%,满足分析要求。
| 表2 梅花鹿瘤胃细菌16S rRNA基因序列分析
Table 2 Sequence analysis of bacteria 16S rRNA gene from the rumen of sika deer
|
107个序列中91个序列(10个OTUs)与已培养细菌的16S rRNA序列相似性≥97%,占总序列的85%。这些已培养菌包括:栖瘤胃普雷沃氏菌(Prevotella ruminicola)(6个OTUs,81.4%总克隆序列)、溶纤维丁酸弧菌(Butyrivibrio fibrisolvens)(1个OTU,0.9%总克隆序列)、Dialister succinatiphilus(1个OTU,0.9%总克隆序列)、Sharpea azabuensis(1个OTU,0.9%总克隆序列)和规则粪球菌(Coprococcus eutactus)(1个OTU,0.9%总克隆序列)。13个序列(10个OTUs,12.2%总克隆序列)与已培养菌16S rRNA序列相似性处于90%~97%,包括:栖瘤胃普雷沃氏菌(2个OTUs,3.7%总克隆序列)、溶纤维丁酸弧菌(2个OTUs,1.8%总克隆序列)、栖牙普雷沃氏菌(Prevotella denticola)(2个OTUs,2.8%总克隆序列)、Dialister succinatiphilus(1个OTU,0.9%总克隆序列)、Solobacterium moorei(1个OTU,0.9%总克隆序列)、Syntrophococcus sucromutans(1个OTU,0.9%总克隆序列)和粪便罗斯拜瑞氏菌(Roseburia faecis)(1个OTU,0.9%总克隆序列)。其余3个序列(2个OTUs,2.8%总克隆序列)与解溶纸梭菌(Clostridium papyrosolvens)相似性<90%。
22个OTUs代表序列与24条相似序列构建系统发育树,结果表明所有序列归类于拟杆菌门和厚壁菌门2大簇(图1)。所有与普雷沃氏菌属(Prevotella spp.)相似序列均聚在拟杆菌门簇,3个相似性低的序列(SDX18、SDX64和SDX68)虽与已培养普雷沃氏菌属聚在一起,但系统发育距离较远,因此可能代表新的普雷沃氏菌属。厚壁菌门簇中,发育关系更为复杂,个别序列并未与其相似序列聚为一类。如粪便罗斯拜瑞氏菌相似序列SDX97(96%)与Syntrophococcus sucromutans在发育关系上更近;溶纤维丁酸弧菌相似序列(SDX56和SDX65)并未与溶纤维丁酸弧菌聚在一起;Dialister succinatiphilus相似序列SDX12(92%)与Solobacterium moorei聚为一类;SDX103和SDX107与解溶纸梭菌聚为一类,但发育距离较远,而且SDX103与牛回肠黏膜未培养细菌序列相似性达到100%,SDX107与黄牛瘤胃中未培养细菌序列相似性为99.6%,因此这些序列可能代表新的未培养的属或种。
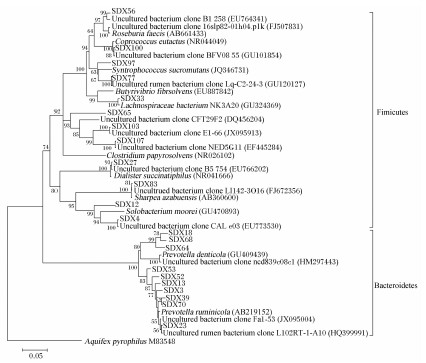 | 图1 梅花鹿瘤胃细菌16S rRNA基因序列系统发育分析
Fig.1 Phylogenetic analysis of bacteria 16S rRNA gene sequences from the rumen of sika deer |
瘤胃是一个含有大量微生物的厌氧生态系统,其中含有细菌1010~1011个/mL、古菌107~109个/mL、原虫104~106个/mL、真菌103~106个/mL和病毒109~1010个/mL[21],这些微生物在降解植物纤维和含毒素植物中发挥着重要作用。在放牧状态下,梅花鹿喜采食单宁含量高的柞树叶。因此,对以柞树叶为主要粗饲料的梅花鹿瘤胃细菌区系进行分析,可为揭示梅花鹿耐受高单宁机制提供依据。
梅花鹿瘤胃细菌区系结果表明,87.9%的16S rRNA基因序列属于与纤维降解密切相关的拟杆菌门,而Pope等[4]等发现挪威驯鹿瘤胃中61%细菌属于拟杆菌门,这可能是因为动物种类或地理环境差异所导致。普雷沃氏菌属是瘤胃中存在数量最为丰富的一类细菌[22, 23, 24],它们可利用多种多糖,并可促进木聚糖的降解[25, 26]。梅花鹿瘤胃中87.9%的16S rRNA基因序列与普雷沃氏菌属相似,而且明显比奶牛(68.5%和42%~60%)[24, 27]、山羊(19.7%)[22]和羊驼(40%)[10]瘤胃中普雷沃氏菌属比例高,这表明普雷沃氏菌属在梅花鹿瘤胃发酵中发挥重要作用。尽管有研究认为,饲粮中添加精饲料可能会导致普雷沃氏菌属在瘤胃中数量增加[28, 29],但梅花鹿瘤胃中普雷沃氏菌属的种类和数量更可能是梅花鹿与瘤胃内微生物互相选择、共同进化的结果。
梅花鹿瘤胃中存在溶纤维丁酸弧菌,但未发现牛、羊瘤胃中常见的纤维素降解菌,如瘤胃球菌属(Ruminococcus spp.)和琥珀酸丝状杆菌(Fibrobacter succinogenes)。McSweeney等[30]发现山羊采食含有单宁的危地马拉朱缨花(Calliandra calothyrsus)后,瘤胃中瘤胃球菌属和琥珀酸丝状杆菌数量减少,这说明单宁能够影响瘤胃中纤维素降解菌的种类。因此,本研究中未检测到瘤胃球菌属和琥珀酸丝状杆菌可能是因为柞树叶中存在的单宁与瘤胃中细菌细胞壁或细胞结合性胞外酶发生反应,细菌可利用养分减少,生长受到抑制。而Jones等[31]报道,栖瘤胃普雷沃氏菌和梭菌属(Clostridium spp.)细菌对缩合单宁具有耐受性,同时McSweeney等[32]发现同一种中不同菌株型对单宁表现出不同的耐受性。另外,由于普雷沃氏菌属具有很高的遗传多样性[22],因此,我们推测梅花鹿对单宁耐受性可能与瘤胃中大量存在的普雷沃氏菌属有关。
序列分析结果表明梅花鹿瘤胃中存在Dialister succinatiphilus、Solobacterium moorei、Syntrophococcus sucromutans、粪便罗斯拜瑞氏菌、规则粪球菌和Sharpea azabuensis,这些细菌也发现于其他反刍动物瘤胃中[23]。王梦芝等[33]认为瘤胃罗斯拜瑞氏菌属(Roseburia sp.)的丁酰辅酶A(CoA):乙酰CoA转移酶可转化乙酸产生丁酸,形成菌种间的交互饲喂,保证瘤胃发酵的流畅性。粪球菌属(Coprococcus sp.)具有降解间苯三酚的功能[34]。考虑到单宁是一种酚类聚合物,所以,梅花鹿瘤胃中粪球菌属也可能参与降解单宁的过程。有研究报道,单宁可以减少反刍动物甲烷排放量[35, 36]。因此,这些细菌可能在梅花鹿瘤胃中具有其他重要功能。因此,研究不同粗饲料对梅花鹿瘤胃中细菌多样性的影响,将有助于理解梅花鹿对单宁的耐受机制。
① 本研究对以富含单宁的柞树叶为主要粗饲料来源的梅花鹿瘤胃细菌多样性进行了初步分析,普雷沃氏菌属是梅花鹿瘤胃内优势细菌,但普雷沃氏菌属相似克隆序列比例与其他反刍动物有一定区别,可能是梅花鹿与瘤胃微生物互相选择、共同进化的结果。
② 常见的纤维素降解菌未检测到可能与饲料中单宁含量高有关。
| [1] | 李国华,秦荣前,梁凤锡.梅花鹿采食草原地区野生植物的调查[J]. 中药材,1986(1):12-14,16. ( 1) 1)
|
| [2] | 杨镒峰,魏海军,李光玉,等.不同单宁水平日粮对育成前期雄性梅花鹿生长和血液生化指标的影响[J]. 特产研究,2010(2):25-29. ( 1) 1)
|
| [3] | HIURA T,HASHIDOKO Y,KOBAYASHI Y,et al.Effective degradation of tannic acid by immobilized rumen microbes of a sika deer (Cervus nippon yesoensis) in winter[J]. Animal Feed Science and Technology,2010,155(1):1-8. ( 1) 1)
|
| [4] | POPE P B,MACKENZIE A K,GREGOR I,et al.Metagenomics of the svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci[J]. PLoS One,2012,7(6):e38571. ( 2) 2)
|
| [5] | SUNDSET M A,EDWARDS J E,CHENG Y F,et al.Molecular diversity of the rumen microbiome of Norwegian reindeer on natural summer pasture[J]. Microbial Ecology,2009,57(2):335-348. ( 1) 1)
|
| [6] | SUNDSET M A,EDWARDS J E,CHENG Y F,et al.Rumen microbial diversity in Svalbard reindeer,with particular emphasis on methanogenic archaea[J]. FEMS Microbiology Ecology,2009,70(3):553-562. ( 1) 1)
|
| [7] | SHI P J,MENG K,ZHOU Z G,et al.The host species affects the microbial community in the goat rumen[J]. Letters in Applied Microbiology,2008,46(1):132-135. ( 1) 1)
|
| [8] | AN D D,DONG X Z,DONG Z Y.Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses[J]. Anaerobe,2005,11(4):207-215. ( 1) 1)
|
| [9] | LENG J,CHENG Y M,ZHANG C Y,et al.Molecular diversity of bacteria in Yunnan yellow cattle (Bos taurs) from Nujiang region,China[J]. Molecular Biology Reports,2012,39(2):1181-1192. ( 1) 1)
|
| [10] | PEI C X,LIU Q A,DONG C S,et al.Diversity and abundance of the bacterial 16S rRNA gene sequences in forestomach of alpacas (Lama pacos) and sheep (Ovis aries)[J]. Anaerobe,2010,16(4):426-432. ( 2) 2)
|
| [11] | YANG S,MA S,CHEN J,et al.Bacterial diversity in the rumen of Gayals (Bos frontalis),Swamp buffaloes (Bubalus bubalis) and Holstein cow as revealed by cloned 16S rRNA gene sequences[J]. Molecular Biology Reports,2010,37(4):2063-2073. ( 1) 1)
|
| [12] | LAMONTAGNE M G,MICHEL F C,HOLDEN P A,et al.Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis[J]. Journal of Microbiological Methods,2002,49(3):255-264. ( 1) 1)
|
| [13] | 郑刚,陈己任,胡博文,等.基于DGGE分析的大鼠粪便及肠道细菌DNA提取方法研究[J]. 食品科学,2011,32(17):215-218. ( 1) 1)
|
| [14] | LANE D J.16S/23S rRNA sequencing[M]. New York:Wiley,1991:115-175. ( 1) 1)
|
| [15] | HUBER T,FAULKNER G,HUGENHOLTZ P.Bellerophon:a program to detect chimeric sequences in multiple sequence alignments[J]. Bioinformatics,2004,20(14):2317-2319. ( 1) 1)
|
| [16] | COLE J R,WANG Q,CARDENAS E,et al.The ribosomal database project:improved alignments and new tools for rRNA analysis[J]. Nucleic Acids Research,2009,37:D141-D145. ( 1) 1)
|
| [17] | SCHLOSS P D,WESTCOTT S L,RYABIN T,et al.Introducing mothur:open-source,platform-independent,community-supported software for describing and comparing microbial communities[J]. Applied and Environmental Microbiology,2009,75(23):7537-7541. ( 1) 1)
|
| [18] | JANDA J M,ABBOTT S L.Bacterial identification for publication:when is enough enough?[J]. Journal of Clinical Microbiology,2002,40(6):1887-1891. ( 1) 1)
|
| [19] | ALTSCHUL S F,MADDEN T L,SCHAFFER A A,et al.Gapped BLAST and PSI-BLAST:a new generation of protein database search programs[J]. Nucleic Acids Research,1997,25(17):3389-3402. ( 1) 1)
|
| [20] | TAMURA K,PETERSON D,PETERSON N,et al.MEGA5:molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods[J]. Molecular Biology and Evolution,2011,28(10):2731-2739. ( 1) 1)
|
| [21] | WRIGHT A D G,KLIEVE A V.Does the complexity of the rumen microbial ecology preclude methane mitigation?[J]. Animal Feed Science and Technology,2011,23:248-253. ( 1) 1)
|
| [22] | BEKELE A Z,KOIKE S,KOBAYASHI Y.Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis[J]. FEMS Microbiology Letters,2010,305(1):49-57. ( 3) 3)
|
| [23] | KIM M,MORRISON M,YU Z.Status of the phylogenetic diversity census of ruminal microbiomes[J]. FEMS Microbiology Ecology,2011,76(1):49-63. ( 2) 2)
|
| [24] | WU S,BALDWIN R L,LI W,et al.The bacterial community composition of the bovine rumen detected using pyrosequencing of 16S rRNA genes[J]. Metagenomics,2012,1:1-11. ( 2) 2)
|
| [25] | KRAUSE D O,DENMAN S E,MACKIE R I,et al.Opportunities to improve fiber degradation in the rumen:microbiology,ecology,and genomics[J]. FEMS Microbiology Reviews,2003,27(5):663-693. ( 1) 1)
|
| [26] | MATSUI H,OGATA K,TAJIMA K,et al.Phenotypic characterization of polysaccharidases produced by four Prevotella type strains[J]. Current Microbiology,2000,41(1):45-49. ( 1) 1)
|
| [27] | STEVENSON D M,WEIMER P J.Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR[J]. Applied Microbiology and Biotechnology,2007,75(1):165-174. ( 1) 1)
|
| [28] | FERNANDO S C,PURVIS H T,NAJAR F Z,et al.Rumen microbial population dynamics during adaptation to a high-grain diet[J]. Applied and Environmental Microbiology,2010,76(22):7482-7490. ( 1) 1)
|
| [29] | SADET-BOURGETEAU S,MARTIN C,MORGAVI D P.Bacterial diversity dynamics in rumen epithelium of wethers fed forage and mixed concentrate forage diets[J]. Veterinary Microbiology,2010,146(1/2):98-104. ( 1) 1)
|
| [30] | MCSWEENEY C S,PALMER B,KENNEDY P M.Effect of Calliandra tannins on rumen microbial function[J]. Animal Production in Australia,1998,22:289. ( 1) 1)
|
| [31] | JONES G A,MCALLISTER T A,MUIR A D,et al.Effects of Sainfoin (Onobrychis viciifolia Scop.) condensed tannins on growth and proteolysis by four strains of ruminal bacteria[J]. Applied and Environmental Microbiology,1994,60(4):1374-1378. ( 1) 1)
|
| [32] | MCSWEENEY C S,PALMER B,MCNEILL D M,et al.Microbial interactions with tannins:nutritional consequences for ruminants[J]. Animal Feed Science and Technology,2001,91(1/2):83-93. ( 1) 1)
|
| [33] | 王梦芝,王洪荣,徐爱秋,等.徐淮白山羊瘤胃细菌和原虫的类群结构研究[J]. 中国农业科学,2009,42(8):2915-2922. ( 1) 1)
|
| [34] | PATEL T R,JURE K G,JONES G A.Catabolism of phloroglucinol by the rumen anaerobe Coprococcus[J]. Applied and Environmental Microbiology,1981,42(6):1010-1017. ( 1) 1)
|
| [35] | LIU H,VADDELLA V,ZHOU D.Effects of chestnut tannins and coconut oil on growth performance,methane emission,ruminal fermentation,and microbial populations in sheep[J]. Journal of Dairy Science,2011,94(12):6069-6077. ( 1) 1)
|
| [36] | TAN H Y,SIEO C C,ABDULLAH N,et al.Effects of condensed tannins from Leucaena on methane production,rumen fermentation and populations of methanogens and protozoa in vitro[J]. Animal Feed Science and Technology,2011,169(3/4):185-193. ( 1) 1)
|




