自1983年Bernier等[7]从枯草芽孢杆菌(Bacillus subtilis)中分离得到木聚糖酶基因,到目前为止,国内外已报道了300余种不同菌株来源的木聚糖酶基因,其中近百种基因被克隆和表达在合适的宿主中[2, 8]。不同来源的木聚糖酶,酶学性质差别较大(表1)[9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25]。细菌木聚糖酶的分子质量在22.5~60.0 ku,最适温度范围为40~75 ℃,最适pH范围为6.0~8.0,其中以7.0居多;热稳定性在40 ℃附近较好,pH稳定性范围为4.5~11.0,主要集中于6.0~9.0。一些放线菌也能产生木聚糖酶,如橄榄绿链霉菌A1(Streptomyces olivaceoviridis A1)[15]和耐盐高温双歧菌YIM 90462T(Thermobifida halotolerans YIM 90462T)[16],其酶分子质量分别为20.8和34.0 ku,最适温度分别为60和90 ℃,最适pH分别为5.2和9.0;热稳定性在60 ℃附近较好,pH稳定性范围分别为4.0~8.8和7.0~8.0。真菌木聚糖酶的酶分子质量在20.0~31.6 ku,最适温度主要集中于50 ℃(50~65 ℃),最适pH范围为3.0~6.0;热稳定性范围为40~60 ℃,平均在50 ℃左右,pH稳定性范围为2.5~10.0,主要集中于3.5~7.5。在木聚糖酶产量方面,真菌与细菌相比具有明显的优势[26];各类产木聚糖酶微生物里都有木聚糖酶活性较高的代表,如细菌里的中度嗜盐菌AX2000(Bacillus alcalophilus AX2000)[12], 它产生的木聚糖酶活性高达25 000 U/mg;放线菌里的橄榄绿链霉菌A1[15],它产生的木聚糖酶活性高达15 000 U/mL;真菌里的青霉菌Pol6(Penicillium occitanis Pol6)[23]和黑曲霉BCC14405(Aspergillus niger BCC14405)[21],它们产生的木聚糖酶活性分别为8 549.85和8 007 U/mg。
| 表1 不同微生物木聚糖酶基因的克隆表达及酶学特性
Table 1 Cloning and expression of different microbial xylanase genes and their enzymology characterization
|
随着分子生物学技术的发展,越来越多的产木聚糖酶微生物被发现,然而环境中还有很多微生物资源不能通过纯培养技术分离得到,近年来发展起来的宏基因组技术解决了这一难题,它通过免培养技术,研究生境中全部微生物遗传物质的总和,挖掘未培养微生物的基因资源[27],在木聚糖酶基因的开发上显示出了巨大的潜力。表2[28, 29, 30, 31, 32, 33, 34, 35, 36, 37]列举了2008—2013年间以环境样本构建宏基因组文库筛选到的木聚糖酶,样品来源于反刍动物瘤胃[29, 31, 32, 34, 35, 36]、土壤[28, 33, 37]和堆肥[30, 37],其中以反刍动物瘤胃居多。常用的文库载体是质粒BAC[29, 32]和FOS[30, 33, 34, 35, 36],宿主是大肠杆菌[28, 29, 30, 31, 32, 33, 34, 35, 36, 37]。文库平均插入片段在5.5~54.5 kb之间,筛选到的克隆数大都在10 000以上,木聚糖酶阳性率最高可达0.14%。
| 表2 利用宏基因组文库筛选到的木聚糖酶
Table 2 Xylanases screening from the metagenomic library
|
蛋白质工程技术,包含蛋白质的理性设计和非理性设计2种方法。传统的理性设计需要预先知道蛋白质的结构、活性位点、催化机制等信息,在清楚结构与功能的前提下,定点突变改变蛋白质分子中个别氨基酸残基,产生新性状的蛋白质[38]。然而,在实际应用中,蛋白质的结构信息很难获取,结构与功能的关系异常复杂,因此理性设计具有很强的限制性[39]。蛋白质的非理性设计,即体外定向进化,它模拟达尔文的自然进化论,利用基因的突变和重组,从体外改造酶的基因,产生基因多样性,并结合定向的筛选最终获得预期性质的进化酶,因它不需要预先知道蛋白质的三维结构信息,弥补了理性设计的不足[40]。它主要包括突变文库的构建、功能表达和文库筛选(选择)3个步骤[41],其核心是突变文库的构建和文库筛选(图1)。
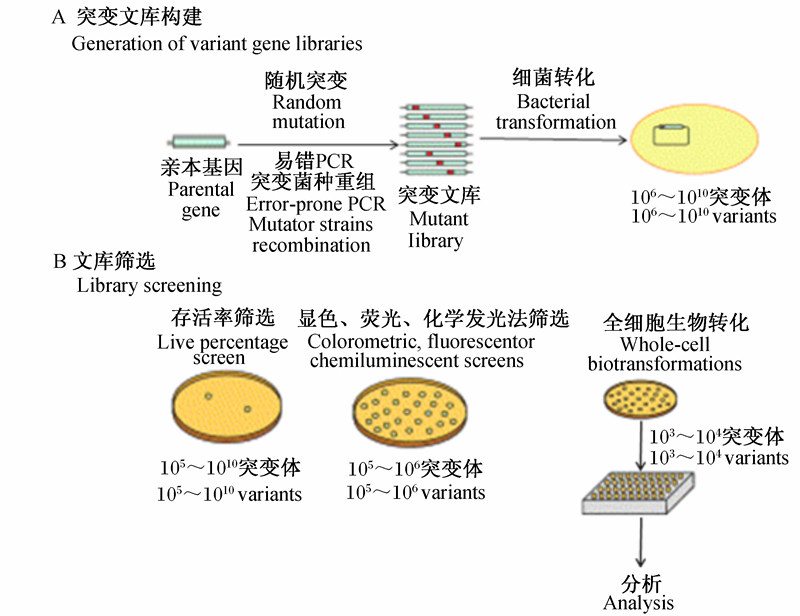 | 图1 酶定向进化的突变文库构建(A)和高通量文库筛选(B)策略 Fig.1 Strategies for the directed evolution of enzymes involving generation of variant gene libraries (A) and high-throughput screening of libraries (B)[42] |
突变文库的库容及多样性是酶分子体外定向进化的基础。构建突变文库的方法有很多,如易错PCR(error-prone PCR)、DNA重排(DNA shuffling),以及基于DNA重排的原理,围绕基因片段重组这一思想产生的交错延伸法(staggered extension process,StEP)、随机引物体外重组(random-priming in vitro recombination,RPR)、退火低核苷酸基因重排(degenerate oligonucleotide gene shuffling,DOGS)、外显子重组(exon shuffling)、酵母增强组合文库(combinatorial libraries enhanced by recombination in yeast,CLERY)、随机片段交换法(random insertional-deletional strand exchange mutagenesis,RAISE)等[38, 40],常用的是易错PCR和DNA重排(表3)[43, 44, 45, 46, 47, 48, 49, 50]。
| 表3 定向进化技术成功改良的微生物木聚糖酶
Table 3 Successfully optimized microbial xylanases by directed evolution
|
易错PCR是定向进化最早采用的一种建库方法,由Leung等[51]提出,后经Cadwell等[52]改良,其原理是在体外扩增目的基因时改变PCR的条件,使碱基产生错配,导致目的基因随机突变。Stephens等[43]用易错PCR产生基因突变,改良1个来自疏棉状嗜热丝孢菌(Thermomyces lanuginosus)的耐热木聚糖酶基因XynA,以提高木聚糖酶热稳定性,其最优突变体酶2B7-10发生1处点突变(Y58F),该突变使其在80 ℃孵育60 min还能保持71%的活性,远高于亲本酶XynA。McHunu等[44]利用易错PCR提高另一个来自疏棉状嗜热丝孢菌(Thermomyces lanuginosus)的木聚糖酶的耐碱性,其突变体酶在60 ℃,pH 10.0的碱性条件下孵育60 min,还能保持84%的活性,而亲本酶同样处理后仅剩22%的活性。Wang等[45]利用易错PCR提高1株杂合木聚糖酶ATx的催化活性,1处氨基酸替换(L49P)使突变体酶的催化活性提高。易错PCR的原理简单,操作简便,对亲本基因的限制条件不多,而且可以和其他突变方法结合使用,因此应用十分广泛,但该方法属于无性突变,遗传只发生在单一分子内部,一般适用于较小的基因片段(<800 bp)[40]。
随着人们对酶的进化期望越来越大,定向进化技术也在不断的成熟和发展。1994年,Stemmer[53, 54]提出DNA重排(DNA shuffling)并成功运用,为体外定向进化技术的飞跃做出了巨大贡献。其原理是用脱氧核糖核酸酶Ⅰ(DNaseⅠ)切割一组含有不同点突变的基因片段,产生不同大小的随机片段,这些片段再经重新组合、扩增形成全长的基因,实现不同基因片段的重组。其优势是可以有效积累有益突变,排除有害和中性突变,同时也能实现蛋白质多种特性的共进化[55]。在木聚糖酶基因定向进化研究上,也有不少应用该技术的例子。Xia等[46]利用DNA重排改良1个来自变铅青链霉菌(Streptomyces lividans)的木聚糖酶B的热稳定性和耐碱性,得到的最优突变体酶在70 ℃可以维持活性时长达6 h,而亲本酶处理3 min后就丧失了50%的活性,此外,突变体酶在pH 9.0的条件下稳定性增加,这些都显示了突变体酶在热稳定性和耐碱性上的极大优势。
DNA重排的前提是存在1组含有不同点突变的基因片段,因而常将DNA重排和易错PCR等进化方法结合使用。Miyazaki等[47]将易错PCR、饱和诱变、DNA重排3种方法结合改良1个来自枯草芽孢杆菌的木聚糖酶的热稳定性,最终得到1个含有3个氨基酸替换的优势突变体Xylst,该突变使其热稳定性显著增加,在60 ℃条件下,突变体酶活性可维持2 h,然而亲本型5 min内就失去了活性。Zhang等[48]利用易错PCR和基于DNA重排的家族重排(family shuffling)技术,与蛋白质半理性设计相结合改良1个来源于嗜热脂肪芽孢杆菌(Geobacillus stearothermophilus)的木聚糖酶XT6的热稳定性,稳定性最佳的突变体酶含有13个氨基酸的替换,该替换导致其半衰期是亲本型的52倍,最适温度从77 ℃上升至87 ℃,催化效率提高90%。
木聚糖酶基因突变文库构建之后,确定一个高通量、高选择性、高灵敏度的筛选方法是快速成功地从庞大的突变库中筛选到目的产物的重要保证[56]。目前常用的筛选方法有平板筛选和基于荧光或显色反应的筛选2种。其中平板筛选最为简便,针对木聚糖酶的平板筛选有RBB-xylan法和刚果红染色法,它是基于木聚糖酶的催化活性和底物分解前后性质的改变,在固体平板中加入木聚糖底物,将克隆点种在相应的选择平板上,宿主菌表达木聚糖酶水解木聚糖形成透明圈,根据透明圈的有无、大小,初步确定是否阳性克隆或优势突变体。但这种筛选局限于底物特异性、酶活性等突变方向的筛选,并且在筛选酶活高的突变体时,也只能作为初步的筛选。对于酶的最适pH、最适温度、稳定性等突变方向,仍需要以可测定的酶促反应结果来筛选,即基于荧光或显色反应的方法,该方法也用于复筛酶活提高的突变体,常用的如3,5-二硝基水杨酸(3,5-dinitrosalicylic acid,DNS)法,它的原理是DNS与木聚糖酶水解木聚糖后产生的还原糖发生氧化还原反应,产物在煮沸条件下显棕红色,且在一定范围内颜色深浅与还原糖含量成比例关系,利用比色法测定还原糖含量,以达到筛选突变体的目的。该方法需要结合96孔板、酶标仪等设备以提高筛选效率[57]。
不同生物来源的木聚糖酶,其酶学性质差别较大,各类产木聚糖酶微生物里都有酶活较高的代表。宏基因组技术在开发环境木聚糖酶基因上显示了巨大的潜力。随着体外定向进化技术的进一步发展和完善,酶的突变、重组、筛选等过程进一步改进,木聚糖酶基因的改良将得到更快的发展。宏基因组技术和体外定向进化技术相结合必将加速木聚糖酶基因的开发和产业化应用,推动半纤维类生物能源的利用。
| [1] | DEUTSCHMANN R,DEKKER R F H.From plant biomass to bio-based chemicals:latest developments in xylan research[J]. Biotechnology Advances,2012,30(6):1627-1640. ( 1) 1)
|
| [2] | 张世敏,刘寅,刘新育,等.木聚糖酶基因研究进展[J]. 微生物学杂志,2006,26(4):61-67. ( 2) 2)
|
| [3] | 朱崇淼,毛胜勇,孙云章,等.产木聚糖酶厌氧真菌菌株筛选及产酶培养条件研究[J]. 微生物学通报,2004,31(3):11-15. ( 1) 1)
|
| [4] | 杨培龙,姚斌.饲料用酶制剂的研究进展与趋势[J]. 生物工程学报,2009,25(12):1844-1851. ( 1) 1)
|
| [5] | 怀文辉,何秀萍,郭文洁,等.微生物木聚糖降解酶研究进展及应用前景[J]. 微生物学通报,2000,27(2):137-139. ( 2) 2)
|
| [6] | 方洛云,邹晓庭,许梓荣.木聚糖酶基因的分子生物学与基因工程[J]. 畜禽业,2002(2):2-3. ( 1) 1)
|
| [7] | BERNIER R,Jr,DRIGUEZ H,DESROCHERS M.Molecular cloning of a Bacillus subtilis xylanase gene in Escherichia coli[J]. Gene,1983,26(1):59-65. ( 1) 1)
|
| [8] | 岳晓禹,贺小营,牛天贵,等.木聚糖酶的研究进展[J]. 酿酒科技,2007(4):113-115,120. ( 1) 1)
|
| [9] | YEASMIN S,KIM C H,PARK H J,et al.Cell surface display of cellulase activity-free xylanase enzyme on Saccharomyces Cerevisiae EBY100[J]. Applied Biochemistry and Biotechnology,2011,164(3):294-304. ( 1) 1)
|
| [10] | BAI W Q,XUE Y F,ZHOU C,et al.Cloning,expression and characterization of a novel salt-tolerant xylanase from Bacillus sp. SN5[J]. Biotechnology Letters,2012,34(11):2093-2099. ( 1) 1)
|
| [11] | CANAKCI S,CEVHER Z,INAN K,et al.Cloning,purification and characterization of an alkali-stable endoxylanase from thermophilic Geobacillus sp. 71[J]. World Journal of Microbiology & Biotechnology,2012,28(5):1981-1988. ( 1) 1)
|
| [12] | LEE D S,LEE K H,CHO E J,et al.Characterization and pH-dependent substrate specificity of alkalophilic xylanase from Bacillus alcalophilus[J]. Journal of Industrial Microbiology & Biotechnology,2012,39(10):1465-1475. ( 2) 2)
|
| [13] | WANG S Y,HU W,LIN X Y,et al.A novel cold-active xylanase from the cellulolytic myxobacterium Sorangium cellulosum So9733-1:gene cloning,expression,and enzymatic characterization[J]. Applied Microbiology and Biotechnology,2012,93(4):1503-1512. ( 1) 1)
|
| [14] | CORRÊA J M,GRACIANO L,ABRAHÃO J,et al.Expression and characterization of a GH39 β-xylosidase Ⅱ from Caulobacter crescentus[J]. Applied Biochemistry and Biotechnology,2012,168(8):2218-2229. ( 1) 1)
|
| [15] | WANG Y R,ZHANG H L,HE Y Z,et al.Characterization,gene cloning,and expression of a novel xylanase XYNB from Streptomyces olivaceoviridis A1[J]. Aquaculture,2007,267(1/2/3/4):328-334. ( 2) 2)
|
| [16] | ZHANG F,CHEN J J,REN W Z,et al.Cloning,expression,and characterization of an alkaline thermostable GH11 xylanase from Thermobifida halotolerans YIM 90462(T)[J]. Journal of Industrial Microbiology & Biotechnology,2012,39(8):1109-1116. ( 2) 2)
|
| [17] | ZHOU C Y,BAI J Y,DENG S S,et al.Cloning of a xylanase gene from Aspergillus usamii and itsexpression in Escherichia coli[J]. Bioresource Technology,2008,99(4):831-838. ( 1) 1)
|
| [18] | HE J,YU B,ZHANG K Y,et al.Expression of a Trichoderma reesei β-xylanase gene in Escherichia coli and activity of the enzyme on fiber-bound substrates[J]. Protein Expression and Purification,2009,67(1):1-6. ( 1) 1)
|
| [19] | WAKIYAMA M,TANAKA H,YOSHIHARA K,et al.Purification and properties of family-10 endo-1,4-β-xylanase from Penicillium citrinum and structural organization of encoding gene[J]. Journal of Bioscience and Bioengineering,2008,105(4):367-374. ( 1) 1)
|
| [20] | PARACHIN N S,SIQUEIRA S,FARIA F P D,et al.Xylanases from Cryptococcus flavus isolate I-11:Enzymatic profile,isolation and heterologous expression of CfXYN1 in Saccharomyces cerevisiae[J]. Journal of Molecular Catalysis B:Enzymatic,2009,59(1/2/3):52-57. ( 1) 1)
|
| [21] | RUANGLEK V,SRIPRANG R,RATANAOHAN N,et al.Cloning,expression,characterization,and high cell-density production of recombinant endo-1,4-β-xylanase from Aspergillus niger in Pichia pastoris[J]. Enzyme and Microbial Technology,2007,41(1/2):19-25. ( 2) 2)
|
| [22] | ZHAO N,GUO R F,YU H W,et al.Expression and characterization of a thermostable xylanase gene xynA from a themophilic fungus in Pichia pastoris[J]. Agricultural Sciences in China,2011,10(3):343-350. ( 1) 1)
|
| [23] | DRISS D,BHIRI F,GHORBEL R,et al.Cloning and constitutive expression of His-tagged xylanase GH 11 from Penicillium occitanis Pol6 in Pichia pastoris X33:purification and characterization[J]. Protein Expression and Purification,2012,83(1):8-14. ( 2) 2)
|
| [24] | HMIDA-SAYARI A,TAKTEK S,ELGHARBI F,et al.Biochemical characterization,cloning and molecular modeling of a detergent and organic solvent-stable family 11 xylanase from the newly isolated Aspergillus niger US368 strain[J]. Process Biochemistry,2012,47(12):1839-1847. ( 1) 1)
|
| [25] | FU G H,WANG Y T,WANG D D,et al.Cloning,expression,and characterization of an GHF 11 xylanase from Aspergillus niger XZ-3S[J]. Indian Journal of Microbiology,2012,52(4):682-688. ( 1) 1)
|
| [26] | AHMED S,RIAZ S,JAMIL A.Molecular cloning of fungal xylanases:an overview[J]. Applied Microbiology and Biotechnology,2009,84(1):19-35. ( 1) 1)
|
| [27] | 王佳堃,安培培,刘建新.宏基因组学用于瘤胃微生物代谢的研究进展[J]. 动物营养学报,2010,22(3):527-535. ( 1) 1)
|
| [28] | HU Y,ZHANG G M,LI A Y,et al.Cloning and enzymatic characterization of a xylanase gene from a soil-derived metagenomic library with an efficient approach[J]. Applied Microbiology and Biotechnology,2008,80(5):823-830. ( 4) 4)
|
| [29] | ZHAO S G,WANG J Q,BU D P,et al.Novel glycoside hydrolases identified by screening a Chinese Holstein dairy cow rumen-derived metagenome library[J]. Applied and Environmental Microbiology,2010,76(19):6701-6705. ( 5) 5)
|
| [30] | KWON E J,JEONG Y S,KIM Y H,et al.Construction of a metagenomic library from compost and screening of cellulase-and xylanase-positive clones[J]. Journal of the Korean Society for Applied Biological Chemistry,2010,53(6):702-708. ( 4) 4)
|
| [31] | CHANG L,DING M Z,BAO L,et al.Characterization of a bifunctional xylanase/endoglucanase from yak rumen microorganisms[J]. Applied Microbiology and Biotechnology,2011,90(6):1933-1942. ( 4) 4)
|
| [32] | GONG X,GRUNINIGER R J,FORSTER R J,et al.Biochemical analysis of a highly specific,pH stable xylanase gene identified from a bovine rumen-derivedmetagenomic library[J]. Applied Microbiology and Biotechnology,2013,97(6):2423-2431. ( 5) 5)
|
| [33] | NACKE H,ENGELHAUPT M,BRADY S,et al.Identification and characterization of novel cellulolytic and hemicellulolytic genes and enzymes derived from German grassland soil metagenomes[J]. Biotechnology Letters,2012,34(4):663-675. ( 5) 5)
|
| [34] | WANG J K,SUN Z Y,ZHOU Y,et al.Screening of a xylanase clone from a Fosmid library of rumen microbiota in Hu sheep[J]. Animal Biotechnology,2012,23(3):156-173. ( 5) 5)
|
| [35] | 王佳堃,安培培,陈振明,等.湖羊瘤胃微生物Fosmid文库的构建和分析[J]. 动物营养学报,2010,22(2):341-345. ( 5) 5)
|
| [36] | RASHAMUSE K J,VISSER D F,HENNESSY F,et al.Characterisation of two bifunctional cellulase-xylanase enzymes isolated from a bovine rumen metagenome library[J]. Current Microbiology,2013,66(2):145-151. ( 5) 5)
|
| [37] | VERMA D,KAWARABAYASI Y,MIYAZAKI K,et al.Cloning,expression and characteristics of a novel alkalistable and thermostable xylanase encoding gene (mxyl) retrieved from compost-soil metagenome[J]. PLoS One,2013,8(1):e52459. ( 4) 4)
|
| [38] | 王黎,袁红霞,曾家豫,等.酶分子定向进化的最新研究进展及应用[J]. 甘肃医药,2009,28(1):24-27. ( 2) 2)
|
| [39] | 徐卉芳,张先恩,张用梅.体外分子定向进化研究进展[J]. 生物化学与生物物理进展,2002,29(4):518-522. ( 1) 1)
|
| [40] | 方柏山,郑媛媛.酶体外定向进化(Ⅰ)突变基因文库构建技术及其新进展[J]. 华侨大学学报:自然科学版,2004,25(4):337-342. ( 3) 3)
|
| [41] | BRAKMANN S.Discovery of superior enzymes by directed molecular evolution[J]. ChemBioChem,2001,2(12):865-871. ( 1) 1)
|
| [42] | TURNER N J.Directed evolution of enzymes for applied biocatalysis[J]. Trends in Biotechnology,2003,21(11):474-478. ( 1) 1)
|
| [43] | STEPHENS D E,RUMBOLD K,PERMAUL K,et al.Directed evolution of the thermostable xylanase from Thermomyces lanuginosus[J]. Journal of Biotechnology,2007,127(3):348-354. ( 3) 3)
|
| [44] | MCHUNU N P,SINGH S,PERMAUL K.Expression of an alkalo-tolerant fungal xylanase enhanced by directed evolution in Pichia pastoris and Escherichia coli[J]. Journal of Biotechnology,2009,141(1/2):26-30. ( 3) 3)
|
| [45] | WANG Q,ZHAO L L,SUN J Y,et al.Enhancing catalytic activity of a hybrid xylanase through single substitution of Leu to Pro near the active site[J]. World Journal of Microbiology and Biotechnology,2012,28(3):929-935. ( 3) 3)
|
| [46] | XIA T,WANG Q.Directed evolution of Streptomyces lividans xylanase B toward enhanced thermal and alkaline pH stability[J]. World Journal of Microbiology and Biotechnology,2009,25(1):93-100. ( 3) 3)
|
| [47] | MIYAZAKI K,TAKENOUCHI M,KONDO H,et al.Thermal stabilization of Bacillus subtilis family-11 xylanase by directed evolution[J]. The Journal of Biological Chemistry,2006,281(15):10236-10242. ( 3) 3)
|
| [48] | ZHANG Z G,YI Z L,PEI X Q,et al.Improving the thermostability of Geobacillus stearothermophilus xylanase XT6 by directed evolution and site-directed mutagenesis[J]. Bioresource Technology,2010,101(23):9272-9278. ( 3) 3)
|
| [49] | CHEN Y L,TANG T Y,CHENG K J.Directed evolution to produce an alkalophilic variant from a Neocallimastix patriciarum xylanase[J]. Canadian Journal of Microbiology,2001,47(12):1088-1094. ( 2) 2)
|
| [50] | TREVIZANO L M,VENTORIN R Z,DE REZENDE S T,et al.Thermostability improvement of Orpinomyces sp. xylanase by directed evolution[J]. Journal of Molecular Catalysis B:Enzymatic,2012,81:12-18. ( 2) 2)
|
| [51] | LEUNG D W,CHEN E,GOEDDEL D V.A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction[J]. Technique,1989,1(1):11-15. ( 1) 1)
|
| [52] | CADWELL R C,JOYCE G F.Randomization of genes by PCR mutagenesis[J]. Genome Research,1992,2(1):28-33. ( 1) 1)
|
| [53] | STEMMER W P C.DNA shuffling by random fragmentation and reassembly:in vitro recombination for molecular evolution[J]. Proceedings of National Academy of Sciences of the United States of America,1994,91(22):10747-10751. ( 1) 1)
|
| [54] | STEMMER W P C.Rapid evolution of a protein in vitro by DNA shuffling[J]. Nature,1994,370:389-391. ( 1) 1)
|
| [55] | 谢晚彬,谢和芳.蛋白质定向进化的研究技术及应用[J]. 中国生物工程杂志,2005,25(S1):16-18. ( 1) 1)
|
| [56] | 方柏山,洪燕,夏启容.酶体外定向进化(Ⅱ)文库筛选的方法及其应用[J]. 华侨大学学报:自然科学版,2005,26(2):113-116. ( 1) 1)
|
| [57] | 王楠,马荣山.酶分子体外定向进化的研究进展[J]. 生物技术通报,2007(2):63-66. ( 1) 1)
|




