2. 中国农业科学院饲料研究所, 国家水产 饲料安全评价基地, 北京 100081;
3. 农业部饲料生物技术重点开放实验室, 北京 100081;
4. 中国海洋大学农业部水产动物营养与饲料重点实验室和海水养殖教育部 重点实验室, 青岛 266003
2. National Aquafeed Safety Assessment Station, Feed Reseach Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China;
3. Key Laboratory of Feed Biotechnology (Ministry of Agriculture), Beijing 100081, China;
4. Key Laboratory of Aquaculture Nutrition and Feed (Ministry of Agriculture) & The Key Laboratory of Mariculture (Ministry of Education), Ocean University of China, Qingdao 266003, China
近年来鱼粉价格不断上涨,鱼粉短缺已成为水产养殖业可持续发展的最大限制性因素,寻找新的优质蛋白质源是亟需解决的问题[1,2]。植物性蛋白质原料由于价格低廉,产量大且供应稳定成为近年来鱼粉替代蛋白质源的研究重点。但植物蛋白质替代鱼粉后会导致肉食性鱼类摄食率出现明显下降,进而降低生长性能[3,4,5,6]。摄食调控着水产养殖动物整个生命周期的生长和能量水平,鱼类的生长与摄食存在明显的相关性[7,8]。植物蛋白质替代鱼粉后对肉食性鱼类摄食的抑制是限制植物蛋白质在肉食性鱼类饲料中有效利用的直接原因[9]。
与哺乳动物类似,鱼类摄食行为受到中枢和外周脑肠肽(ghrelin)-神经肽Y(NPY)和雷帕霉素靶蛋白(mTOR)-NPY途径的调控,但具体机制尚不清楚[9]。mTOR是细胞内关键性的能量感受器,能够感知营养物质、生长因子、激素信号的变化,参与调控蛋白质及脂肪合成[10,11]。mTOR在机体能量平衡和代谢中具有不可替代的作用,在中枢神经系统中作用于神经通路调控摄食和体重,在外周器官主要影响葡萄糖和脂肪代谢[12]。核糖体蛋白S6激酶beta-1(S6K1)可能是mTOR的下游效应器,与翻译的起始和延伸有关,其磷酸化与否可以作为mTOR在细胞和组织中活性启动与否的一种标志[13]。NPY是由36个氨基酸构成的胰多肽,由NPY前体(NPY precursor)基因翻译后经蛋白酶水解加工产生[14]。NPY广泛分布在鱼类的中枢和外周神经系统中[15],是强有效的促食欲神经肽,研究证明外源注射NPY能够促进草鱼(Ctenopharyngodon idellus)[16]和罗非鱼(Oreochromis sp.)[17]的摄食,而日本鳗鲡(Anguilla japonica)[18]、大西洋鳕鱼[19]和美洲拟鲽(Pseudopleuronectes americanus)[20]在饥饿状态下下丘脑中NPY precursor mRNA的表达量显著升高。ghrelin又称为生长激素促分泌素,由28个氨基酸组成,是由脑肠肽前体(preproghrelin)基因翻译后辛酰化第3个氨基酸形成的具有生物活性的多肽[21]。ghrelin同样是促食欲神经肽,外源注射ghrelin能够促进斑点叉尾 (Ictalurus punctatus)[22]的摄食,饥饿则能诱导欧鲈(Dicentrarchus labrax)[23]胃中preproghrelin表达量的增加。
花鲈(Lateolabrax japonicus)是我国重要的肉食性海水养殖品种。张志勇[24]报道,在必需氨基酸平衡模式下使用混合植物蛋白质替代全部鱼粉,会导致花鲈的摄食率下降73%,而血浆中NPY水平显著升高,这说明花鲈即便处于饥饿状态,仍然拒食全植物蛋白质饲料,因此有必要从摄食调控的角度探索高植物蛋白质饲料造成花鲈摄食抑制的分子机制。本试验采用反转录聚合酶链式反应(RT-PCR)和cDNA末端快速扩增(RACE)技术花鲈摄食调控相关因子cDNA全长序列,为进一步研究高植物蛋白质饲料造成花鲈摄食抑制的分子调控机制,提高肉食性鱼类对植物蛋白质的利用及人工选育高植物蛋白质耐受鲈鱼新品种奠定基础。
1 材料与方法 1.1 试验材料试验所用花鲈购自山东潍坊环海水产良种场。
1.2 引物设计与合成根据NCBI GenBank中已知其他物种的mTOR、S6K1、NPY precursor和preproghrelin相应的保守氨基酸序列设计简并引物克隆花鲈各个基因的核心片段。再根据所得目的片段设计用于mTOR的接头特异性引物和各个基因的3′RACE、5′RACE PCR扩增特异性引物。本试验所用的引物均由生工生物工程(上海)有限公司合成。所有引物序列及退火温度(Tm)见表1。
1.3 总RNA提取使用RNAiso Plus(总RNA提取试剂盒,TaKaRa)分别提取花鲈肝脏(用于克隆mTOR和S6K1)、下丘脑(用于克隆NPY precursor)和胃(用于克隆preproghrelin)中总RNA。采用Power-WaveTM XS2全波长酶标仪(BioTek)对所提取的总RNA进行定量。
1.4 核心片段克隆参照PrimeScript Ⅱ 1st Strand cDNA Synthesis Kit (TaKaRa)说明书合成cDNA第1链,保守片段聚合酶链式反应(PCR)的体系包括合成的cDNA第1链1 uL,TaKaRa Ex Taq (5 U/μL)0.125 μL,10×Ex Taq Buffer (Mg2+ Plus)2.5 μL,dNTP Mixture (2.5 mmol/L)2 μL,上游引物(10 μmol/L)1 μL,下游引物(10 μmol/L)1 μL,加灭菌蒸馏水至25 μL。PCR扩增条件:94 ℃,30 s;Tm,30 s;72 ℃,1 min;共35个循环,72 ℃,5 min。琼脂糖凝胶变性电泳检验PCR产物,切割回收目的片段并参照MiniBEST Agarose Gel DNA Extraction Kit Version 3.0 (TaKaRa)说明书进行纯化。在PCR仪上,16 ℃连接,冰浴中转化,经蓝白斑筛选后进行测序(北京华大基因服务公司)。
1.5 3′-RACE扩增采用3′-Full RACE Core Set with PrimeScriptTM RTase (TaKaRa)进行3′-RACE扩增,操作步骤参照说明书。1)Outer PCR反应。反应体系:cDNA 2 μL;1×cDNA Dilution Buffer Ⅱ 3 μL,Gene Specific Outer Primer (10 μmol/L)1 μL,3′-RACE Outer Primer (10 μmol/L)1 μL,10×LA PCR Buffer Ⅱ (Mg2+ Free)2 μL,MgCl2 (25 mmol/L)1.5 μL,TaKaRa LA Taq (5 U/μL)0.125 μL,加dH2O至25 μL。反应条件:94 ℃,3 min;94 ℃,30 s;Tm,30 s;72 ℃,1 min (mTOR 3 min);共35循环,72 ℃,5 min。②Inner PCR反应。反应体系:1st PCR产物(稀释1 000倍) 0.5 μL,dNTP Mixture (2.5 mmol/L)4 μL,10×LA PCR Buffer Ⅱ(Mg2+ Free)2.5 μL,MgCl2 (25 mmol/L)2.5 μL,TaKaRa LA Taq (5 U/μL)0.25 μL,Gene Specific Inner Primer (10 μmol/L)1 μL,3′-RACE Inner Primer (10 μmol/L)1 μL,加dH2O至25 μL。反应条件:94 ℃,3 min;94 ℃,30 s;Tm,30 s;72 ℃,1 min (mTOR 3 min);共30循环,72 ℃,5 min。
| 表1 本试验中所用到的引物序列及相应的退火温度 Table 1 Primer sequence and Tm used in this experiment |
参照SMARTerTM RACE cDNA Amplification Kit(Clontech)说明书进行5′-RACE扩增。反应体系:PCR-Grade Water 17.25 μL,10×Advantage 2 PCR Buffer 2.5 μL,dNTP Mix(10 mmol/L)0.5 μL,50×Advantage 2 Polymerase Mix 0.5 μL,cDNA 1.25 μL,UPM (10×)2.5 μL,GSP 0.5 μL。Touch-Down PCR反应程序:5个循环94 ℃,30 s;72 ℃,2 min;5个循环94 ℃,30 s;70 ℃,30 s;72 ℃,2 min;25个循环94 ℃,30 s; 68 ℃,30 s;72 ℃,2 min。
1.7 序列分析、同源性比对及系统进化树构建将测序所得的核心片段、3′末端和5′末端序列进行拼接即获得各个基因的全长。将最终获得的结果与NCBI GenBank中已知其他物种序列做同源性分析以确定是否是花鲈摄食调控因子的蛋白质,再用DNAMAN(version 5.2.2)对目的基因进行cDNA序列分析。根据GenBank中已知物种的摄食调控因子的蛋白质序列用MegAlign进行多序列比对和一致性数值的计算。选择邻接法(neighbor-joining,NJ)构建系统进化树。
2 结果与分析 2.1 mTOR、S6K1、NPY precursor和preproghrelin cDNA全长序列图1显示,花鲈mTOR cDNA全长序列为8 296 bp,其中5′末端非翻译区共有210 bp,中间的序列为开放阅读框,共有7 551 bp,其余的则为3′末端非翻译区,共有535 bp。花鲈mTOR的开放阅读框共编码2 516个氨基酸,其蛋白质分子质量预测为286.14 ku,等电点为6.52。花鲈mTOR cDNA全长序列在GenBank中的登录号为KJ746670。
 | 开放阅读框7 551 bp,编码2 516个氨基酸。起始密码子和终止密码子用黑体表示。 The 7 551 bp ORF encodes a protein of 2 516 amino acids in length. The start and stop codons are indicated by boldface.图1 花鲈mTOR cDNA全长序列和预测的氨基酸序列(GenBank登录号KJ746670) Fig. 1 mTOR cDNA full length sequence and deduced amino acid sequence of Lateolabrax japonicus (GenBank accession No. KJ746670) |
图2显示,花鲈S6K1 cDNA全长序列为2 232 bp,其中5′末端非翻译区共有126 bp,中间的序列为开放阅读框,共有1 539 bp,其余的则为3′末端非翻译区,共有567 bp。花鲈S6K1的开放阅读框共编码512个氨基酸,其蛋白质分子质量预测为56.87 ku,等电点为5.88。花鲈S6K1 cDNA全长序列在GenBank中的登录号为KJ746671。
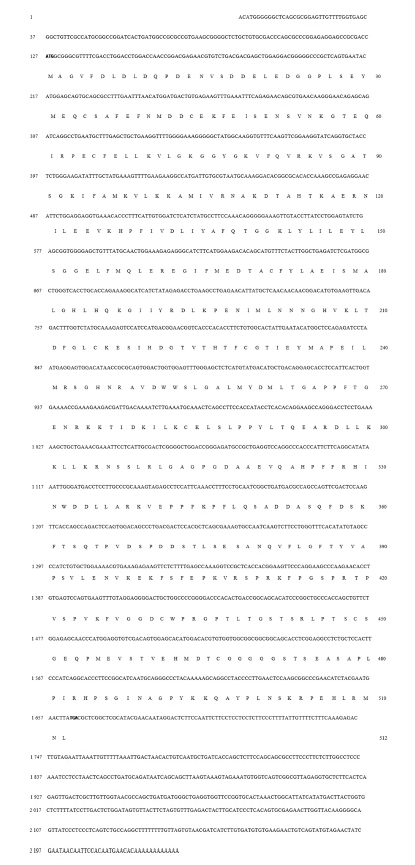 | 开放阅读框1 539 bp,编码512个氨基酸。起始密码子和终止密码子用黑体表示。 The 1 539 bp ORF encodes a protein of 512 amino acids in length. The start and stop codons are indicated by boldface.图2 鲈S6K1 cDNA全长序列和预测的氨基酸序列(GenBank登录号KJ746671) Fig. 2 2 S6K1 cDNA full length sequence and deduced amino acid sequence of Lateolabrax japonicus (GenBank accession No. KJ746671) |
图3显示,花鲈NPY precursor cDNA全长序列为676 bp,其中5′末端非翻译区共有66 bp,中间的序列为开放阅读框,共有300 bp,其余的则为3′末端非翻译区,共有310 bp。花鲈NPY precursor的开放阅读框共编码99个氨基酸,其蛋白质分子质量预测为11.26 ku,等电点为5.26。花鲈NPY precursor cDNA全长序列在GenBank中的登录号为KJ850326。
 | 开放阅读框300 bp,编码99个氨基酸。起始密码子和终止密码子用黑体表示。 The 300 bp ORF encodes a protein of 99 amino acids in length. The start and stop codons are indicated by boldface.图3 花鲈NPY precursor cDNA全长序列和预测的氨基酸序列(GenBank登录号KJ850326) Fig. 3 NPY precursor cDNA full length sequence and deduced amino acid sequence of Lateolabrax japonicus (GenBank accession No. KJ850326) |
图4显示,花鲈preproghrelin cDNA全长序列为1 201 bp,其中5′末端非翻译区共有140 bp,中间的序列为开放阅读框,共有324 bp,其余的则为3′末端非翻译区,共有737 bp。花鲈preproghrelin的开放阅读框共编码107个氨基酸,其蛋白质分子质量预测为12.03 ku,等电点为5.39。花鲈preproghrelin cDNA全长序列在GenBank中的登录号为KJ850327。
 | 开放阅读框324 bp,编码107个氨基酸。起始密码子和终止密码子用黑体表示。 The 324 bp ORF encodes a protein of 107 amino acids in length. The start and stop codons are indicated by boldface.图4 花鲈preproghrelin cDNA全长序列和预测的氨基酸序列(GenBank登录号KJ850327) Fig. 4 Preproghrelin cDNA full length sequence and deduced amino acid sequence of Lateolabrax japonicus (GenBank accession No. KJ850327) |
图5为花鲈mTOR氨基酸序列与其他多个物种同源性分析结果,结果显示花鲈mTOR氨基酸序列与斑马鱼(Danio rerio)同源性最高,为98.0%,与斑点叉尾 、鲤鱼(Cyprinus carpio)、人(Homo sapiens)、小鼠(Mus musculus)、绵羊(Ovis aries)、眼镜王蛇(Ophiophagus hannah)的同源性分别为96.1%、97.9%、90.6%、90.7%、90.5%、62.0%。
 | 比对所用到的物种和GenBank登录号如下:斑马鱼,NP_001070679.2;小鼠,NP_064393.2;人,NP_004949.1;斑点叉尾鮰,AHH40208.1;绵羊,NP_001138927.1;鲤鱼,FJ899680.1;眼镜王蛇,ETE69347.1。“.”表示与顶行的花鲈氨基酸序列相同的位点。 Used species with GenBank accession No. for alignment as follows: Danio rerio, NP_001070679.2; Mus musculus, NP_064393.2; Homo sapiens, NP_004949.1; Ictalurus punctatus, AHH40208.1; Ovis aries, NP_001138927.1; Cyprinus carpio, FJ899680.1; Ophiophagus hannah, ETE69347.1. Sites with an amino acid identical to that on the top line (Lateolabrax japonicus) are indicated with a dot.图5 花鲈与其他物种mTOR氨基酸序列比对结果Fig. 5 Alignment results of mTOR amino acid sequences between Lateolabrax japonicus and others species |
图6为花鲈S6K1氨基酸序列与其他多个物种同源性分析结果,结果显示花鲈S6K1氨基酸序列与斑马鱼同源性最高,为84.9%,与米氏叶吻银鲛(Callorhinchus milii)、家兔(Oryctolagus cuniculus)、响尾蛇(Crotalus horridus)、褐家鼠(Rattus norvegicus)、原鸡(Gallus gallus)、爪蟾(Xenopus tropicalis)的同源性分别为80.4%、80.9%、81.9%、80.7%、82.3%、80.9%。
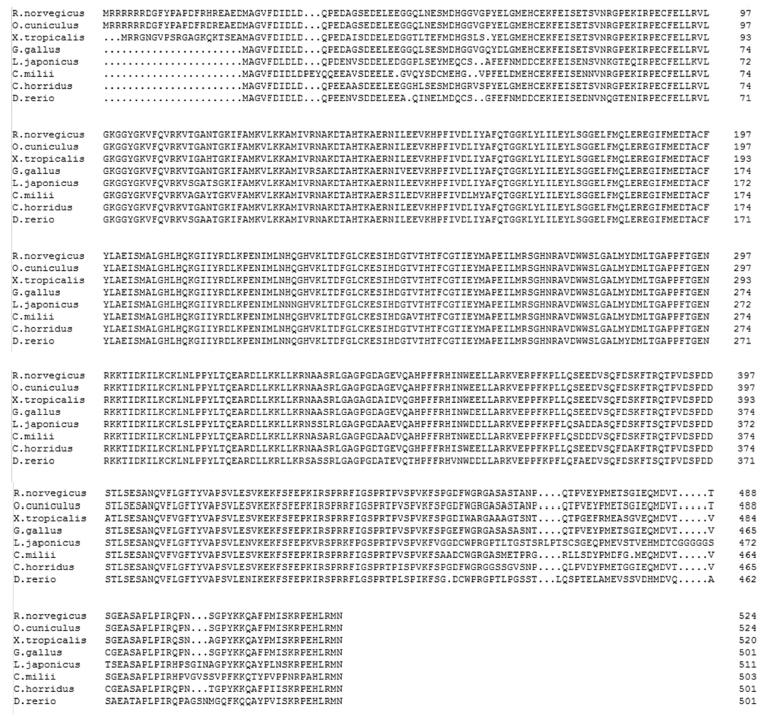 | 比对所用到的物种和GenBank登录号如下:大鼠,NP_114191.1;家兔,NP_001095160.1;非洲爪蟾,NP_001034817.1;原鸡,NP_001025892.1;叶吻银鲛,AFO98445.1;响尾蛇,JAA95741.1;斑马鱼,NP_998241.1。“.”表示与顶行的花鲈氨基酸序列相同的位点。 Used species with GenBank accession No. for alignment as follows: Rattus norvegicus, NP_114191.1; Oryctolagus cuniculus, NP_001095160.1; Xenopus (Silurana) tropicalis, NP_001034817.1; Gallus gallus, NP_001025892.1; Callorhinchus milii, AFO98445.1; Crotalus horridus, JAA95741.1; Danio rerio, NP_998241.1. Sites with an amino acid identical to that on the top line (Lateolabrax japonicus) are indicated with a dot.图6 花鲈与其他物种S6K1氨基酸序列比对结果 Fig. 6 Alignment results of S6K1 amino acid sequences between Lateolabrax japonicus and others species |
图7为花鲈NPY precursor氨基酸序列与其他多个物种同源性分析结果,结果显示花鲈NPY precursor氨基酸序列与军曹鱼(Rachycentron canadum)同源性最高,为99.0%,与欧鲈、斑马鱼、团头鲂(Megalobrama amblycephala)、人、家兔、小鼠、原鸡、爪蟾的同源性分别为98.0%、59.4%、61.5%、58.8%、57.7%、60.8%、59.8%、53.6%。
 | 比对所用到的物种和GenBank登录号如下:斑马鱼,AAI62068.1;非洲爪蟾,NP_001161232.1;人,AAA59944.1;军曹鱼,AGN03939.1;小鼠,AAH43012.1;团头鲂,AFO68117.1;家兔,BAG82945.1;原鸡,AAA48991.1;欧鲈,CAB64932.1。“.”表示与顶行的花鲈氨基酸序列相同的位点。黑色阴影是花鲈成熟NPY序列。 Used species with GenBank accession No. for alignment as follows: Danio rerio, AAI62068.1; Xenopus laevis, NP_001161232.1; Homo sapiens, AAA59944.1; Rachycentron canadum, AGN03939.1; Mus musculus, AAH43012.1; Megalobrama amblycephala, AFO68117.1; Oryctolagus cuniculus, BAG82945.1; Gallus gallus, AAA48991.1; Dicentrarchus labrax, CAB64932.1. Sites with an amino acid identical to that on the top line (Lateolabrax japonicus) are indicated with a dot. The black boxes with white letters represent the 36 amino acid peptide sequence of mature NPY.图7 花鲈与其他物种NPY precursor氨基酸序列比对结果Fig. 7 Alignment results of NPY precursor amino acid sequences between Lateolabrax japonicus and others species |
图8为花鲈preproghrelin氨基酸序列与其他多个物种同源性分析结果,结果显示花鲈preproghrelin氨基酸序列与欧鲈的同源性最高,为85%,与罗非鱼、黑棘鲷(Acanthopagrus schlegelii)、人、绵羊、小鼠、爪蟾、原鸡的同源性分别为74.8%、81.3%、21.5%、24.3%、23.4%、19.6%、25.2%。
 | 比对所用到的物种和GenBank登录号如下:小鼠,BAB19046.1;罗非鱼,BAC65152.1;绵羊,ABC00742.1;原鸡,AAP56234.1;黑棘鲷,AAV65509.1;人,BAA89371.1;非洲爪蟾,NP_001267573.1;欧鲈,ABG49130.1。“.”表示与顶行的花鲈氨基酸序列相同的位点。黑色阴影是成熟ghrelin序列。 Used species with GenBank accession No. for alignment as follows: Mus musculus, BAB19046.1; Oreochromis niloticus, BAC65152.1; Ovis aries, ABC00742.1; Gallus gallus, AAP56234.1; Acanthopagrus schlegelii, AAV65509.1; Homo sapiens, BAA89371.1; Xenopus laevis, NP_001267573.1; Dicentrarchus labrax, ABG49130.1. Sites with an amino acid identical to that on the top line (Lateolabrax japonicus) are indicated with a dot. The black boxes with white letters represent the amino acid peptide sequence of mature ghrelin.图8 花鲈与其他物种preproghrelin氨基酸序列比对结果 Fig. 8 Alignment results of preproghrelin amino acid sequences between Lateolabrax japonicus and others species |
图9为花鲈mTOR氨基酸序列系统进化树,如图所示,花鲈mTOR以bootstrap值为99的支持度与斑马鱼和鲤鱼合并为一支,然后与斑点叉尾 合并为鱼类分支;眼镜王蛇以100的支持度与小鼠、人、绵羊等哺乳动物合并为另外一支。
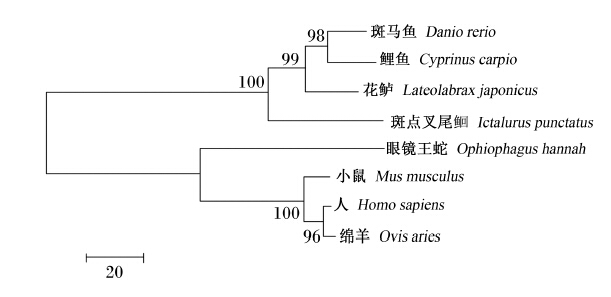 | 系统进化树构建所用的物种和GenBank登录号如下: 斑马鱼,NP_001070679.2;小鼠,NP_064393.2;人,NP_004949.1;斑点叉尾,AHH40208.1;绵羊,NP_001138927.1;鲤鱼,FJ899680.1;眼镜王蛇,ETE69347.1。分支上的数字代表bootstrap值。 Used species with GenBank accession No. for phylogenetic tree construction as follows: Danio rerio, NP_001070679.2; Mus musculus, NP_064393.2; Homo sapiens, NP_004949.1; Ictalurus punctatus, AHH40208.1; Ovis aries, NP_001138927.1; Cyprinus carpio, FJ899680.1; Ophiophagus hannah, ETE69347.1.The number in the branch is the bootstrap value.图9 花鲈和其他物种mTOR氨基酸序列系统进化树聚类分析Fig. 9 Cluster analysis of phylogenetic tree of mTOR amino acid sequence between Lateolabrax japonicus and other species |
图10为花鲈S6K1氨基酸序列系统进化树,如图所示,花鲈S6K1以bootstrap值为100的支持度与斑马鱼合并为一支;哺乳类以84的支持度先与原鸡合并,然后以88的支持度与响尾蛇合并,再以99的支持度与热带爪蟾合并,最后与叶吻银鲛合并为另外一支。
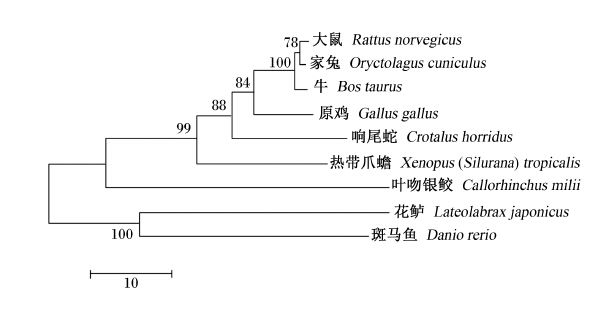 | 系统进化树构建所用的物种和GenBank登录号如下:大鼠,NP_114191.1;家兔,NP_001095160.1;牛,NP_991385.1;斑马鱼,NP_998241.1;热带爪蟾,NP_001034817.1;原鸡,NP_001025892.1;响尾蛇,JAA95741.1;叶吻银鲛,AFO98445.1。分支上的数字代表bootstrap值。 Used species with GenBank accession No. for phylogenetic tree construction as follows: Rattus norvegicus, NP_114191.1; Oryctolagus cuniculus, NP_001095160.1; Bos taurus, NP_991385.1; Danio rerio, NP_998241.1; Xenopus (Silurana) tropicalis, NP_001034817.1; Gallus gallus, NP_001025892.1; Crotalus horridus, JAA95741.1; Callorhinchus milii, AFO98445.1. The number in the branch is the bootstrap value.图10 花鲈和其他物种S6K1氨基酸序列系统进化树聚类分析 Fig. 10 Cluster analysis of phylogenetic tree of S6K1 amino acid sequence between Lateolabrax japonicus and other species |
图11为NPY precursor氨基酸序列系统进化树,如图所示,花鲈NPY precursor以bootstrap值为47的支持度与军曹鱼合并,然后以100的支持度与欧鲈合并为一支,再以89的支持度与斑马鱼、团头鲂合并为鱼类分支;非洲爪蟾以49的支持度先与原鸡合并,然后以69的支持度与哺乳类合并,最后与石纹电鳐合并为另外一支。
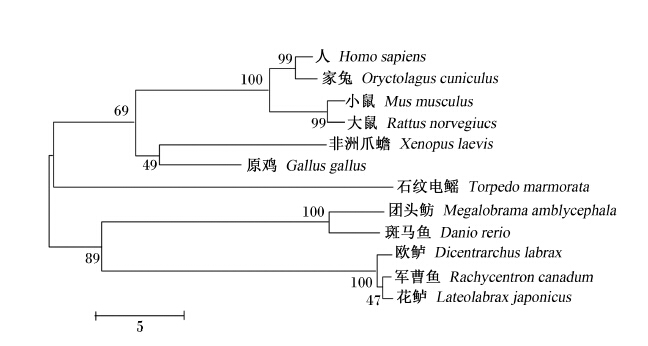 | 系统进化树构建所用的物种和GenBank登录号如下:非洲爪蟾,L07413.1;人,NM_000905.3;军曹鱼,KC284716.1;小鼠,AF273768.1;团头鲂,JQ301475.1;家兔,NM_001160286.1;原鸡,NM_205473.1;石纹电鳐,M87296.1;欧鲈,AJ005378.2;斑马鱼,NM_131074.2;大鼠,NM_012614.2。分支上的数字代表bootstrap值。 Used species with GenBank accession No. for phylogenetic tree construction as follows: Xenopus laevis, L07413.1; Homo sapiens, NM_000905.3; Rachycentron canadum, KC284716.1; Mus musculus, AF273768.1; Megalobrama amblycephala, JQ301475.1; Oryctolagus cuniculus, NM_001160286.1; Gallus gallus, NM_205473.1; Torpedo marmorata, M87296.1; Dicentrarchus labrax, AJ005378.2; Danio rerio, NM_131074.2; Rattus norvegicus, NM_012614.2. The number in the branch is the bootstrap value.图11 花鲈和其他物种NPY precursor氨基酸序列系统进化树聚类分析 Fig. 11 Cluster analysis of phylogenetic tree of NPY precursor amino acid sequence between Lateolabrax japonicus and other species |
图12为preproghrelin氨基酸序列系统进化树,如图所示,花鲈preproghrelin以bootstrap值为70的支持度与欧鲈合并,然后以93的支持度与黑棘鲷合并,最后以100的支持度与罗非鱼合并为鱼类分支;原鸡先以89的支持度与哺乳类合并,然后与非洲爪蟾合并为另外一支。
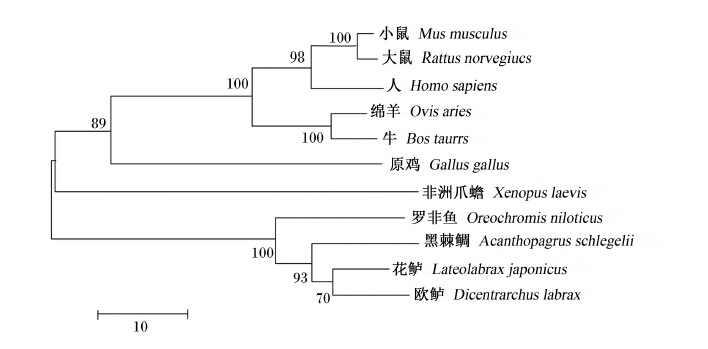 | 系统进化树构建所用的物种和GenBank登录号如下:小鼠,AB035701.1;罗非鱼,AB104859.1;绵羊,DQ294307.1;原鸡,AY299454.1;黑棘鲷,AY643808.1;人,AB029434.1;非洲爪蟾,AB669180.1;欧鲈,DQ665912.1;大鼠,AB029433.1;牛,AF350329.1。分支上的数字代表bootstrap值。 Used species with GenBank accession No. for phylogenetic tree construction as follows: Mus musculus, AB035701.1; Oreochromis niloticus, AB104859.1; Ovis aries, DQ294307.1; Gallus gallus, AY299454.1; Acanthopagrus schlegelii, AY643808.1; Homo sapiens, AB029434.1; Xenopus laevis, AB669180.1; Dicentrarchus labrax, DQ665912.1; Rattus norvegicus, AB029433.1; Bos taurus, AF350329.1. The number in the branch is the bootstrap value.图12 花鲈和其他物种preproghrelin氨基酸序列系统进化树聚类分析Fig. 12 Cluster analysis of phylogenetic tree of preproghrelin amino acid sequence between Lateolabrax japonicus and other species |
mTOR是生长因子和营养信号的整合器。本试验克隆到的花鲈mTOR是一种磷脂酰肌醇激酶,属于磷脂酰肌醇三激酶相关蛋白激酶(phosphoinositide 3-kinase-related protein kinase,PIKK)家族中的蛋白激酶。雷帕霉素结合区域、雷帕霉素催化区域及碳端区域构成了mTOR高度保守区。1 982 aa~2 081 aa是雷帕霉素结合区域,该区域可形成alpha螺旋结构;2 120 aa~2 398 aa是雷帕霉素的催化区域,也是其他酶如丝氨酸/苏氨酸/酪氨酸激酶、氨基糖苷磷酸转移酶、胆碱激酶及RIO激酶的催化区域;2 484 aa~2 516 aa构成了FATC区域,该区域主要作用是维持mTOR的结构稳定性。另外,838 aa~1 008 aa是功能未知 区域,其普遍存在于真核生物中,花鲈mTOR的结构与其他物种相似[25,26,27]。通过多序列比对发现花鲈mTOR氨基酸序列与其他鱼类的相似性大于96%,与哺乳动物的相似性大于90%,说明其在进化中具有较高的保守性。目前尚没有关于mTOR对鱼类摄食调控影响的研究报道。
核糖体蛋白S6激酶(ribosomal protein S6 kinases,S6Ks)是mTOR合成蛋白质的下游信号通路[28]。在摄食调控方面,影响摄食的因子直接作用于mTOR或其下游效应器(可能是S6K1),来调控摄食[9]。S6K1主要参与调控蛋白质合成、细胞生长和细胞增殖。小鼠在饥饿时,S6K1通过解除对下丘脑中神经肽的抑制和减少瘦素对下丘脑的敏感度,有效促进摄食,增加体重[29]。通过多序列比对发现花鲈S6K1氨基酸序列与斑马鱼的相似性很高(84.9%),与其他脊椎动物的相似性均大于80%,由此可见S6K1在进化过程中具有很高的保守性,系统进化树分析结果也表明其与鱼类的亲缘关系更近。
NPY是已知最有效的促食因子[30,31],广泛分布在鱼类中央神经系统中,其中下丘脑中丰度最高[30],在金鱼(Carassius auratus)、大西洋鲑(Salmo salar)、斑点叉尾 中已证实NPY能剂量依赖性增加鱼类摄食[32,33,34,35]。但不同鱼类的NPY在摄食调控网络中的作用机制会有所差别,可能与鱼类食性分化有关[16,36]。花鲈NPY precursor开放阅读框编码99个氨基酸,包括由前28个氨基酸组成的信号肽、紧接着是36个氨基酸构成的成熟NPY区域及碳端区域。花鲈成熟NPY氨基酸序列与团头鲂、斑马鱼的完全相同,与人、非洲爪蟾、小鼠、军曹鱼、家兔及欧鲈仅1个氨基酸不同,说明成熟NPY在进化中具有高度的保守性,其在摄食调控中的作用可能与其他物种相同。
preproghrelin翻译后进行酰化形成ghrelin[37],ghrelin是外周食欲增强的第1反应信号,具有促进生长激素释放的功能,由胃和肠合成。花鲈ghrelin是由20个氨基酸组成的,其中前4个氨基酸“GSSF”与哺乳动物相同,是其活性中心[38],成熟ghrelin第3位置上的丝氨酸是ghrelin的酰化位点,此位点对于识别ghrelin受体具有重要作用[39]。成熟ghrelin多肽氮端氨基酸序列比碳端保守,可变性小,经多序列比对发现前7个氨基酸在脊椎动物中高度保守,是ghrelin生物活性位点。花鲈ghrelin生物活性位点的高度保守对于维持机体生长和代谢十分重要,其作用机理也许与哺乳动物相同[23,40]。
在哺乳动物中已证明ghrelin和NPY对摄食的促进是通过mTOR/S6K1信号通路。给哺乳动物注射ghrelin或其在饥饿条件下,会诱导mTOR活性的增加[41]。在鼠中已经证实,中枢神经系统中的ghrelin通过磷酸化诱导激活S6K1,mTOR/S6K1活性的增加对于ghrelin发挥其促进摄食作用必不可少[42]。NPY对摄食的促进作用需要通过ghrelin,NPY神经元细胞内钙离子浓度的升高会激活钙离子依赖激酶激酶2(calmodulin-dependent kinase kinase 2,CAMKK2),在CAMKK2的作用下ghrelin激活AMP活化蛋白激酶(AMP-activated protein kinase,AMPK),AMPK磷酸化乙酰辅酶A,活化肉毒碱棕榈酰转移酶1(carnitine palmitoyl transferase 1,CPT1),从而增强NPY的表达[43,44]。AMPK调控的ghrelin促食过程不同于mTOR/S6K1信号通路,在鱼类中两者之间有何差异及2个信号通路对ghrelin的摄食调控作用哪个占主导作用目前研究较少。由高植物蛋白质饲料导致的花鲈厌食状态下各摄食调控因子是如何调控及其与哺乳动物摄食调控机理的异同都需要进一步的研究,本试验成功克隆了花鲈摄食调控相关因子的cDNA全长序列,为下一步的摄食调控机理研究奠定了基础。
4 结 论本试验成功克隆到花鲈摄食调控关键因子mTOR、S6K1、NPY precursor和preproghrelin的cDNA全长序列,经多序列比对发现其与已知物种相应基因的氨基酸序列具有很高的相似性。系统进化树分析显示,花鲈的以上基因均与其他鱼类的亲缘关系较近。
| [1] | NAYLOR R L,GOLDBURG R J,PRIMAVERA J H,et al.Effect of aquaculture on world fish supplies[J]. Nature,2000,405(6790):1017-1024. ( 1) 1)
|
| [2] | HU L,YUN B,XUE M,et al.Effects of fish meal quality and fish meal substitution by animal protein blend on growth performance,flesh quality and liver histology of Japanese sea bass (Lateolabrax japonicus)[J]. Aquaculture,2013,372-375:52-61. ( 1) 1)
|
| [3] | HANSEN A C,ROSENLUND G,KARLSEN Ø,et al.Total replacement of fish meal with plant proteins in diets for Atlantic cod (Gadus morhua L.) Ⅰ—effects on growth and protein retention[J]. Aquaculture,2007,272(1/2/3/4;:599-611. ( 1) 1)
|
| [4] | GÔMEZ-REQUENI P,MINGARRO M,CALDUCH-GINER J A,et al.Protein growth performance,amino acid utilisation and somatotropic axis responsiveness to fish meal replacement by plant protein sources in gilthead sea bream (Sparus aurata;[J]. Aquaculture,2004,232(1/2/3/4):493-510. ( 1) 1)
|
| [5] | KAUSHIK S J,CRAVEDI J P,LALLES J P,et al.Partial or total replacement of fish meal by soybean protein on growth,protein utilization,potential estrogenic or antigenic effects,cholesterolemia and flesh quality in rainbow trout,Oncorhynchus mykiss[J]. Aquaculture,1995,133(3/4):257-274. ( 1) 1)
|
| [6] | SALZE G,MCLEAN E,BATTLE P R,et al.Use of soy protein concentrate and novel ingredients in the total elimination of fish meal and fish oil in diets for juvenile cobia,Rachycentron canadum[J]. Aquaculture,2010,298(3/4):294-299. ( 1) 1)
|
| [7] | 崔奕波.鱼类生物能量学的理论与方法[J]. 水生生物学报,1989,13(4):369-383. ( 1) 1)
|
| [8] | BRETT J R.Physiological energetics[M]//HOARD W S,RANDALL D J,BRETT J R.Fish physiology. New York:Academic Press,1979,8:279-352. ( 1) 1)
|
| [9] | 王嘉,薛敏,吴秀峰,等.鱼类对不同蛋白质源饲料选择性摄食调控机制的研究进展[J]. 动物营养学报,2014,26(4):833-842. ( 3) 3)
|
| [10] | COSTA-MATTIOLI M,MONTEGGIA L M.mTOR complexes in neurodevelopmental and neuropsychiatric disorders[J]. Nature Neuroscience,2013,16(11):1537-1543. ( 1) 1)
|
| [11] | LAPLANTE M,SABATINI D M.mTOR signaling in growth control and disease[J]. Cell,2012,149(2):274-293. ( 1) 1)
|
| [12] | HAISSAGUERRE M,SAUCISSE N,COTA D.Influence of mTOR in energy and metabolic homeostasis[J]. Molecular and Cellular Endocrinology,2014,397(1/2):67-77. ( 1) 1)
|
| [13] | UM S H,FRIGERIO F,WATANABE M,et al.Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity[J]. Nature,2004,431(7005):200-205. ( 1) 1)
|
| [14] | LARHAMMAR D.Evolution of neuropeptide Y,peptide YY and pancreatic polypeptide[J]. Regulatory Peptides,1996,62(1):1-11. ( 1) 1)
|
| [15] | THORSELL A,HEILIG M.Diverse functions of neuropeptide Y revealed using genetically modified animals[J]. Neuropeptides,2002,36(2/3):182-193. ( 1) 1)
|
| [16] | ZHOU Y,LIANG X F,YUAN X C,et al.Neuropeptide Y stimulates food intake and regulates metabolism in grass carp,Ctenopharyngodon idellus[J]. Aquaculture,2013,380381//382/383:52-61. ( 2) 2)
|
| [17] | CARPIO Y,ACOSTA J,MORALES A,et al.Cloning,expression and growth promoting action of Red tilapia (Oreochromis sp.) neuropeptide Y[J]. Peptides,2006,27(4):710-718. ( 1) 1)
|
| [18] | LI S S,ZHAO L P,XIAO L,et al.Structural and functional characterization of neuropeptide Y in a primitive teleost,the Japanese eel(Anguilla japonica)[J]. General and Comparative Endocrinology,2012,179(1;:99-106. ( 1) 1)
|
| [19] | KEHOE A S,VOLKOFF H.Cloning and characterization of neuropeptide Y (NPY) and cocaine and amphetamine regulated transcript (CART) in Atlantic cod (Gadus morhua)[J]. Comparative Biochemistry and Physiology Part A:Molecular & Integrative Physiology,2007,146(3):451-461. ( 1) 1)
|
| [20] | MACDONALD E,VOLKOFF H.Cloning,distribution and effects of season and nutritional status on the expression of neuropeptide Y (NPY),cocaine and amphetamine regulated transcript (CART) and cholecystokinin (CCK) in winter flounder (Pseudopleuronectes americanus) [J]. Hormones and Behavior,2009,56(1):58-65. ( 1) 1)
|
| [21] | KOJIMA M,HOSODA H,DATE Y,et al.Ghrelin is a growth-hormone-releasing acylated peptide from stomach[J]. Nature,1999,402(6762):656-660. ( 1) 1)
|
| [22] | KAIYA H,SMALL B C,BILODEAU A L,et al.Purification,cDNA cloning,and characterization of ghrelin in channel catfish,Ictalurus punctatus[J]. General and Comparative Endocrinology,2005,143(3):201-210. ( 1) 1)
|
| [23] | TEROVA G,RIMOLDI S,BERNARDINI G,et al.Sea bass ghrelin:molecular cloning and mRNA quantification during fasting and refeeding[J]. General and Comparative Endocrinology,2008,155(2):341-351. ( 2) 2)
|
| [24] | 张志勇.花鲈和西伯利亚鲟利用植物蛋白源的差异及GH/IGF-Ⅰ轴调节机制的比较研究[D]. 硕士学位论文.北京:中国农业科学院,2013. ( 1) 1)
|
| [25] | BOSOTTI R,ISACCHI A,SONNHAMMER E L L.FAT:a novel domain in PIK-related kinases[J]. Trends in Biochemical Sciences,2000,25(5):225-227. ( 1) 1)
|
| [26] | DAMES S A,MULET J M,RATHGEB-SZABO K,et al.The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability[J]. The Journal of Biological Chemistry,2005,280(21):20558-20564. ( 1) 1)
|
| [27] | VEVERKA V,CRABBE T,BIRD I,et al.Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor:compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR[J]. Oncogene,2008,27(5):585-595. ( 1) 1)
|
| [28] | FINGAR D C,RICHARDSON C J,TEE A R,et al.mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E[J]. Molecular and Cellular Biology,2004,24(1):200-216. ( 1) 1)
|
| [29] | BLOUET C,ONO H,SCHWARTZ G J.Mediobasal hypothalamic p70 S6 kinase 1 modulates the control of energy homeostasis[J]. Cell Metabolism,2008,8(6):459-467. ( 1) 1)
|
| [30] | HALFORD J C G,COOPER G D,DOVEY T M.The pharmacology of human appetite expression[J]. Current Drug Targets,2004,5(3):221-240. ( 2) 2)
|
| [31] | KALRA S P,DUBE M G,PU S Y,et al.Interacting appetite-regulating pathways in the hypothalamic regulation of body weight[J]. Endocrine Reviews,1999,20(1):68-100. ( 1) 1)
|
| [32] | DE PEDRO N,LÔPEZ-PATIÑO M A,GUIJARRO A I,et al.NPY receptors and opioidergic system are involved in NPY-induced feeding in goldfish[J]. Peptides,2000,21(10):1495-1502. ( 1) 1)
|
| [33] | LÔPEZ-PATIÑO M A,GUIJARRO A I,ISORNA E,et al.Neuropeptide Y has a stimulatory action on feeding behavior in goldfish (Carassius auratus)[J]. European Journal of Pharmacology,1999,377(2/3):147-153. ( 1) 1)
|
| [34] | NARNAWARE Y K,PETER R E.Effects of food deprivation and refeeding on neuropeptide Y (NPY) mRNA levels in goldfish[J]. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology,2001,129(2/3):633-637. ( 1) 1)
|
| [35] | SILVERSTEIN J T,PLISETSKAYA E M.The effects of NPY and insulin on food intake regulation in fish[J]. American Zoologist,2000,40(2):296-308. ( 1) 1)
|
| [36] | LIANG X F,LI G Z,YAO W,et al.Molecular characterization of neuropeptide Y gene in Chinese perch,an acanthomorph fish[J]. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology,2007,148(1):55-64. ( 1) 1)
|
| [37] | KIRSZ K,ZIEBA D A.Ghrelin-mediated appetite regulation in the central nervous system[J]. Peptides,2011,32(11):2256-2264. ( 1) 1)
|
| [38] | BEDNAREK M A,FEIGHNER S D,PONG S S,et al.Structure-function studies on the new growth hormone-releasing peptide,ghrelin:minimal sequence of ghrelin necessary for activation of growth hormone secretagogue receptor 1a[J]. Journal of Medicinal Chemistry,2000,43(23):4370-4376. ( 1) 1)
|
| [39] | MUCCIOLI D G,PAPOTTI M,LOCATELLI V,et al.Binding of 125I-labeled ghrelin to membranes from human hypothalamus and pituitary gland[J]. Journal of Endocrinological Investigation,2001,24(3):RC7-RC9. ( 1) 1)
|
| [40] | VOLKOFF H,CANOSA L F,UNNIAPPAN S,et al.Neuropeptides and the control of food intake in fish[J]. General and Comparative Endocrinology,2005,142(1/2):3-19. ( 1) 1)
|
| [41] | VILLANUEVA E C,MVNZBERG H,COTA D,et al.Complex regulation of mammalian target of rapamycin complex 1 in the basomedial hypothalamus by leptin and nutritional status[J]. Endocrinology,2009,150(10):4541-4551. ( 1) 1)
|
| [42] | STEVANOVIC D,TRAJKOVIC V,MVLLER-LVHLHOFF S,et al.Ghrelin-induced food intake and adiposity depend on central mTORC1/S6K1 signaling[J]. Molecular and Cellular Endocrinology,2013,381(1/2):280-290. ( 1) 1)
|
| [43] | ANDREWS Z B.Central mechanisms involved in the orexigenic actions of ghrelin[J]. Peptides,2011,32(11):2248-2255. ( 1) 1)
|
| [44] | KOHNO D,SONE H,MINOKOSHI Y,et al.Ghrelin raises [Ca2+]<i>i via AMPK in hypothalamic arcuate nucleus NPY neurons[J]. Biochemical and Biophysical Research Communications,2008,366(2):388-392. ( 1) 1)
|




