2. 江苏省海洋生物产业技术协同创新中心, 连云港 222005;
3. 浙江省淡水水产研究所, 湖州 313001
2. Co-Innovation Center of Jiangsu Marine Bio-Industry Technology, Lianyungang 222005, China;
3. Zhejiang Institute of Freshwater Fisheries, Huzhou 313001, China
乙酰辅酶A羧化酶(ACC,EC 6.4.1.2)催化乙酰辅酶A生成丙二酸单酰辅酶A,是脂肪酸从头合成的限速酶[1, 2]。原核生物ACC由3个基因编码的亚基组成,它们分别是生物素羧化酶(BC)、生物素羧基载体蛋白(BCCP)和羧基转移酶(CT)[3]。真核生物ACC由1个基因编码,并具有上述3个结构域。哺乳动物ACC有α和β 2种类型,它们具有不同的组织分布和功能。ACC-α通常称为ACACA或ACC1,主要在脂肪生成组织如肝脏中表达,催化长链脂肪酸的从头合成[4, 5, 6]。ACC-β通常称为ACACB或ACC2,是线粒体酶,主要在心脏和肌肉中表达,其产物丙二酰辅酶A通过抑制长链脂酰辅酶A从胞液向线粒体的转运而调控脂肪酸氧化[7, 8]。在恒温动物中,已克隆出鼠[4]、鸡[5]、人[6]、牛[9]、绵羊[10]和山羊[11]ACC1基因的全长cDNA。人的ACC1基因含有1个7 041 bp的开放阅读框,编码2 346个氨基酸,ACC1蛋白分子质量255.5 ku;ACC2蛋白分子质量为280 ku。这2类ACC都由独立的基因进行编码,ACC1基因在染色体17q12上,ACC2基因在染色体12q23上,两者的总氨基酸序列的相似性达到76%,两者的差别是在N端,ACC2比ACC1多大约140个氨基酸,而ACC2就是通过这些多出的氨基酸序列锚定在线粒体的外膜上。关于鱼类ACC基因的研究较少,已克隆出草鱼ACC1基因全长cDNA序列,该基因主要在草鱼肝脏和脂肪组织中表达[12]。
半滑舌鳎是我国重要的海水养殖鱼类,但近年来其脂肪代谢相关疾病逐年增加,影响商品鱼肉质和成活率。本研究的目的是克隆半滑舌鳎ACC1基因全长cDNA,分析其组织表达,并研究饲料中不同脂肪水平对其表达的影响,探讨该基因在脂肪代谢中的作用,为鱼类脂肪代谢营养调控研究提供基础资料和理论依据。
1 材料与方法 1.1 半滑舌鳎ACC1基因全长cDNA分子克隆 1.1.1 试验用鱼试验用半滑舌鳎购自江苏省赣榆现代渔业示范园区,个体湿重750~1 000 g,在淮海工学院海洋学院240 L水族箱中饲养2周,水温20~23 ℃。
1.1.2 试验引物本试验所用引物见表1。各引物在ACC1 cDNA上的位置见图1。
| 表1 试验所用引物序列及预期产物大小 Table 1 Primer sequences and expected product size for the experiment |
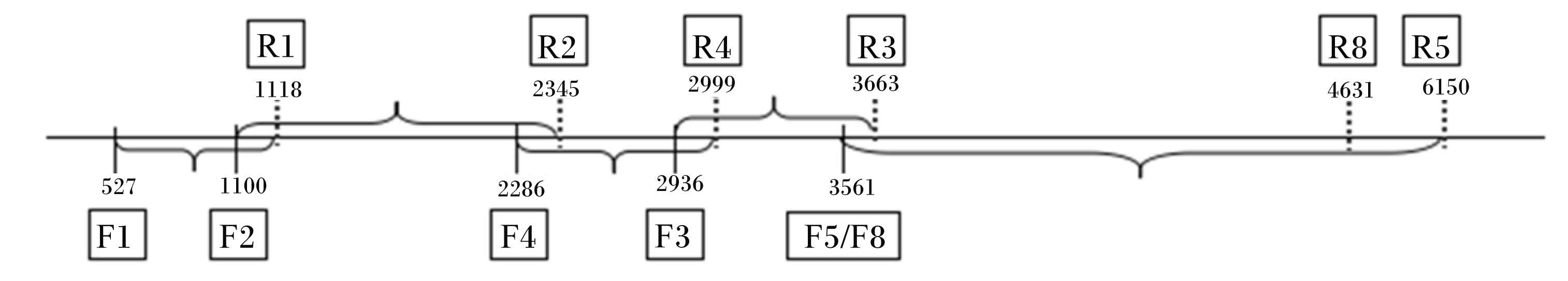 | 图1 各引物在ACC1 cDNA上的位置 Fig. 1 Locations of primers in ACC1 cDNA |
半滑舌鳎活体解剖,快速分离肝脏,液氮研磨,使用QIAGEN公司的RNeasy Lipid Tissue Mini Kit试剂盒,按推荐方法提取肝脏总RNA。1%的琼脂糖凝胶电泳检测RNA质量,核酸定量仪(Gene Quant)检测RNA的浓度后-70 ℃保存备用。
1.1.4 用于核心序列扩增的第1链cDNA反转录采用QIAGEN的QuantiTect Reverse Transcription Kit反转录试剂盒,以Oligo(dT)16AP为引物,按推荐方法去除DNA污染并反转录合成cDNA第1链,稀释10倍备用。
1.1.5 核心序列扩增根据斑马鱼(XM_001919780)ACC1基因序列,设计5对简并引物(表1和图1),PCR扩增ACC1基因5段核心序列,25 μL反应体系含:TaKaRa公司的EmeraldAmp PCR Master Mix 12.5 μL,cDNA第1链1 μL,上、下游引物各0.5 μL,ddH2O 10.5 μL。扩增条件为:94 ℃变性40 s、52 ℃退火40 s、72 ℃延伸60 s,共35个循环;反应前95 ℃预变性5 min;反应后72 ℃充分延伸7 min。1%的琼脂糖凝胶电泳检测,切胶回收,克隆测序。
1.1.6 半滑舌鳎ACC1基因3′cDNA末端快速扩增(RACE)根据ACC1基因核心序列设计3′RACE特异引物(表1)。以cDNA第1链为模版,ACC3-F1和AP为引物进行第1轮PCR扩增,将第1轮的PCR产物稀释10倍为模板,以ACC3-F2和Race3-R为引物进行第2轮扩增。3′RACE的反应体系和反应条件同核心序列,1%的琼脂糖凝胶电泳检测,切取目的条带并回收,克隆测序。
1.1.7 半滑舌鳎ACC1基因5′RACE参考Dieffenbach等[13]的方法快速扩增ACC1基因5′末端,5′RACE扩增使用的引物见表1。首先,以ACC5-R为引物反转录合成cDNA第1链;然后,用RNase H分解cDNA-RNA杂交体中的RNA,乙醇沉淀;再用末端脱氧核苷转移酶在5′末端加poly(A)尾,20 μL反应体系如下:在有沉淀的原管中加入5×Buffer 4 μL,dATP(100 mmol/L)1 μL,末端脱氧核苷转移酶(20 U/μL)1.5 μL,加ddH2O至20 μL。反应条件为:37 ℃温育30 min,70 ℃灭活10 min,用TE缓冲液稀释10倍后备用;最后,以Oligo(dT)16AP和ACC5-R1为引物进行第1轮PCR扩增,产物稀释10倍,用Race3-R与ACC5-R2进行第2轮PCR扩增,PCR反应体系和条件同核心序列。1%琼脂糖凝胶电泳检测,切取目的条带并回收,克隆测序。
1.1.8 序列分析用DNAstar 7.1软件包中的SeqMan对核心序列以及3′和5′末端序列进行组装,得到ACC1基因全长cDNA序列。用EditSeq对序列进行编辑和分析,寻找开放阅读框,并翻译成氨基酸序列。通过http://www.cbs.dtu.dk/services/预测糖基化位点、磷酸化位点;通过http://www.cbs.dtu.dk/services/SignalP/预测信号肽;通过http://www.ebi.ac.uk/interpro/和http://www.cbs.dtu.dk/services/TMHMM/预测跨膜结构域。使用MEGA 6.0采用邻接(neighbor joining,NJ)法构建基于ACC1氨基酸序列的分子系统发育树[14]。
1.2 半滑舌鳎ACC1基因实时荧光定量PCR采用SYBR Green实时荧光定量PCR方法,对ACC1基因表达进行定量研究。
1.2.1 内参基因的克隆与定量引物设计内参基因β2微球蛋白(beta-2 microglobulin,B2M)和核糖体蛋白L13A(ribosomal protein L13a,RPL13A)从GenBank中获得,登录号分别为FJ965562和GH232293;核糖体蛋白L4(ribosomal protein L4,RPL4)和肽基脯氨酰顺反异构酶A(peptidyl-prolyl cis-trans isomerase A,PPIase)采用电子克隆方法获得。内参基因引物见表1。
1.2.2 定量PCR检测体系实时荧光定量PCR反应在StepOnePlus PCR仪(ABI)上进行。采用QIAGEN的QuantiFast SYBR Green PCR Kit定量试剂盒,20 μL的反应体系中包含2×PCR Master Mix 10 μL,25 mmol/L的上、下游引物各0.8 μL和10倍稀释后的cDNA第1链1 μL,ddH2O 7.4 μL。采用2步PCR法进行扩增,首先95 ℃预变性5 min,然后是40个循环,每个循环包括95 ℃变性10 s和60 ℃延伸30 s,循环结束后,从60 ℃缓慢升温到95 ℃,制备熔解曲线。每次反应都设置无模板对照,每个样品设3个技术重复。
1.2.3 半滑舌鳎ACC1基因的组织表达取3尾喂食后6 h的半滑舌鳎活体解剖,快速分离肠道、肝脏、肌肉、卵巢、脾脏、全脑、肾脏、心脏、肠系膜脂肪等组织,分别提取总RNA。采用QIAGEN的QuantiTect Reverse Transcription Kit反转录试剂盒,以Primer Mix为引物,按试剂盒推荐方法去除DNA污染并反转录合成cDNA第1链,稀释10倍用于荧光定量分析,经筛选确定RPL13A、RPL4、PPIase为组织表达的3个内参基因。
1.2.4 饲料脂肪水平对半滑舌鳎肝脏中ACC1基因表达的影响 1.2.4.1 试验饲料共配制4种试验饲料,4种试验饲料的鱼油添加量分别为0(对照)、3.5%、7.0%和10.0%(表2)。制作饲料的鱼粉、鱼油、胆碱等主要原料由淮安通威饲料科技有限公司提供。根据配方,称量所需要的各种原料,充分混匀10 min,加水后揉拌15 min,再利用便携式绞肉机进行压缩制粒。将制粒的饲料均匀洒在白瓷盘内,放入干燥箱内50 ℃烘10 h,室温放置1 h,装袋、编号、密封保存待用。
| 表2 试验饲料组成及营养水平(干物质基础) Table 2 Composition and nutrient levels of experimental diets (DM basis) |
试验在“单循环可控实验生态水槽系统”(大连汇新钛设备开发有限公司生产)内完成,每个水槽体积240 L。整套系统有独立的水处理、温度控制、充气/增氧和紫外消毒等设备。试验用鱼购自江苏省赣榆现代渔业示范园区,共240尾,规格为76.7~87.6 g/尾,随机分为4组,每组3个重复(水槽),每个重复20尾,养殖周期12周,养殖水温20~23 ℃。日投饲2次,投饲量随各水槽内半滑舌鳎的摄食情况及时调整,以30 min内吃完为度。
1.2.4.3 ACC1基因相对表达量测定养殖12周后,每组随机取3尾喂食后6 h的半滑舌鳎提取肝脏总RNA,反转录合成cDNA第1链,采用SYBR Green实时荧光定量PCR方法测定ACC1基因的相对表达量。经筛选确定RPL4和B2M为内参基因。
1.2.5 数据分析将内参基因的扩增效率和循环数(Cq)值数据导入geNorm程序,从而为不同的处理选择不同的内参组合。将目的基因和内参基因的扩增效率和Cq值数据导入qBase Plus程序,从而计算各基因在不同的处理下的相对表达量,结果采用平均值±标准差表示。统计分析采用SPSS 13.0软件,各组数据均通过了正态性和方差齐性检验,因此,组间差异采用单因素方差分析(one-way ANOVA),组间多重比较采用Turkey’s检验,P<0.05表示差异显著。
2 结果与分析 2.1 半滑舌鳎ACC1基因全长cDNA分子特征半滑舌鳎ACC1基因cDNA全长7 811 bp(图2),含1个7 074 bp的开放阅读框,编码2 357个氨基酸,ACC1蛋白计算分子质量为266 ku,等电点6.42(GenBank登录号为KP033455)。此外,由于可变剪接,还发现了另外2个分子质量分别为265和264 ku的半滑舌鳎ACC1的同工型(isoforms)(GenBank登录号分别为KP033456和KP033457),与分子质量为266 ku的ACC1相比,分别少8和15个氨基酸。
 |
*代表终止密码子,灰色碱基代表实时荧光定量PCR和验证可变剪接的引物,灰色氨基酸表示信号肽,保守的磷酸化位点Ser79~Ser81、潜在的ATP结合域Gly316~Gly321、生物素结合位点Val785~Met788及辅酶A结合位点Ser1969~Val1995用氨基酸加框表示。
* represented a termination codon.The grey bases indicated the primers for RT-qPCR and verifying alternative splicing.The grey amion acids indicated signal peptides.Boxed amion acid sequences indicate the conservative phosphorylation sites Ser79 to Ser81,the potential ATP-binding site Gly316 to Gly321,the biotin-binding site Val785 to Met788,and the CoA-binding site Ser1969 to Val1995. 图2 半滑舌鳎ACC1基因全长cDNA核苷酸序列及其编码氨基酸序列 Fig. 2 The nucleotide sequence and deduced amino acid sequences of ACC1 gene full-length cDNA in half-smooth tongue sole (Cynoglossus semilaevis) |
以RPL13A、RPL4、PPIase 3个基因组合为内参基因,采用实时荧光定量PCR方法对半滑舌鳎ACC1 mRNA在肠道、肝脏、肌肉、卵巢、脾脏、全脑、肾脏、心脏、肠系膜脂肪等组织中的表达进行 了研究。结果表明,所有组织中均检测到ACC1 mRNA的表达,肝脏和全脑中的相对表达量显著高于其他组织(P<0.05),其相对表达量分别为2.67和2.53,肠道、卵巢和肠系膜脂肪中相对表达量次之,分别为1.14、0.97和1.10,肾脏中相对表达量最低,仅为0.48(图3)。
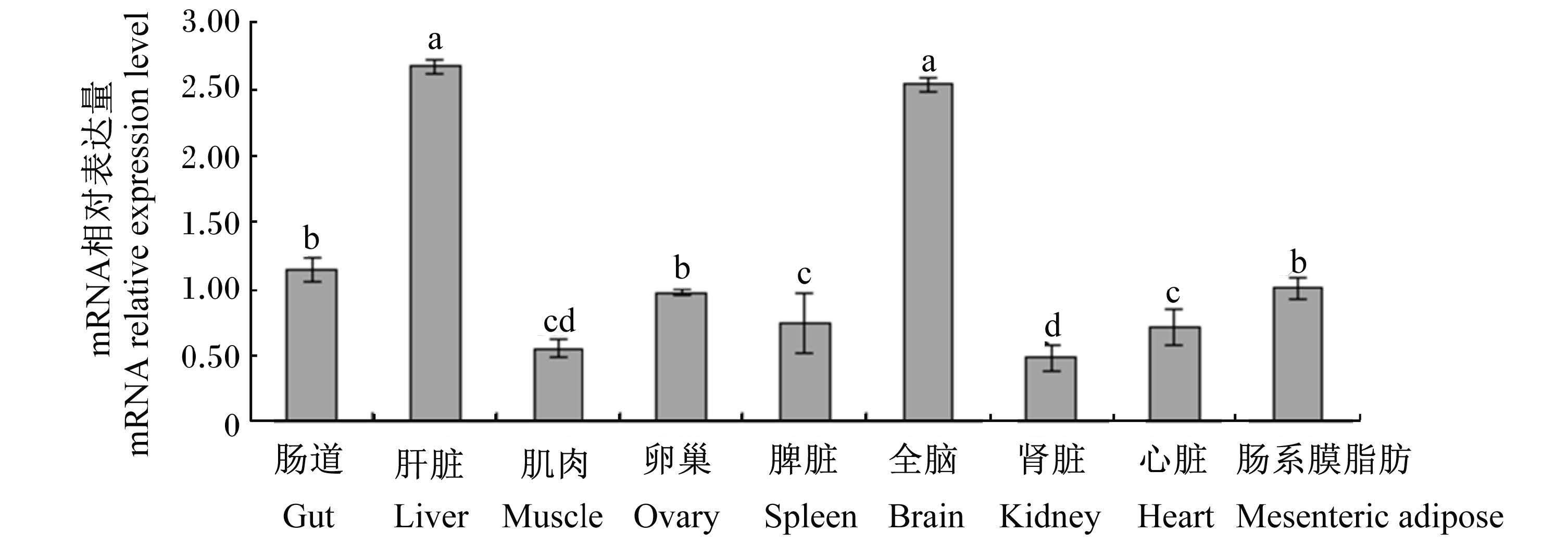 |
数据柱标注不同字母表示差异显著(P<0.05)。图5同。
Value columns with different letters mean significant difference (P<0.05).The same as Fig.5. 图3 半滑舌鳎ACC1基因组织表达分析 Fig. 3 Expression analysis of ACC1 gene in different tissues of half-smooth tongue sole (Cynoglossus semilaevis) |
从GenBank下载了7种鱼类和16种其他脊椎动物ACC1氨基酸序列用于系统发育分析。使用MEGA 6.0软件的Clustal W方法进行氨基酸序列比对,用NJ法构建基于ACC1氨基酸序列的系统发育树,用自展法(bootstrap method)进行系统发生检测,结果如图4所示。
由图4可见,所有鱼类聚为一枝,与其他脊椎动物分开。在鱼类中,同属鲤科的斑马鱼(Danio rerio)(GenBank登录号为XP_009300128和XP_009300133)、草鱼(Ctenopharyngodon idella)(GenBank登录号为ADT82650)、墨西哥脂鲤(Astyanax mexicanus)(GenBank登录号为XP_007245267)聚为一枝,自展值为99%;本研究克隆的半滑舌鳎的3个同工型首先聚为一枝,然后与同属鲈形总目(Percomorpha)的大黄鱼(Laimichthys crocea)(GenBank登录号为XP_010748451)、布氏新灿鲷(Neolamprologus brichardi)(GenBank登录号为XP_006799564)和月光鱼(Xiphophorus maculatus)(GenBank登录号为XP_005803670)聚为一枝,自展值为99%,表明它们的亲缘关系较近。其他动物中,哺乳类、鸟类和爬行类均单独聚为一枝,自展值均为100%,其中鸟类首先和爬行类聚为一枝,然后再与哺乳类聚为一枝,最后与两栖类聚为一枝。在哺乳类中,偶蹄动物牛(Bos taurus)(GenBank登录号为XP_005220033)和山羊(Capra hircus)(GenBank登录号为XP_005693213)聚为一枝;熊科动物大熊猫(Ailuropoda melanoleuca)(GenBank登录号为XP_002924940)和北极熊(Ursus maritimus)(GenBank登录号为XP_00869366)聚为一枝;灵长类动物智人(Homo sapiens)(GenBank登录号为NP_942131和XP_006721916)、黑猩猩(Pan trolodytes)(GenBank登录号为XP_511428)和普通猕猴(Macaca mulatta)(GenBank登录号为NP_001253707)聚为一枝,自展值均为100%。基于ACC1氨基酸序列的系统发育关系与传统分类基本一致。此外,由于ACC1存在多种形式的可变剪接,每种生物的ACC1均有多个同工型,如斑马鱼的同工型多达13个,但每种生物的同工型都首先聚在一起,表明这些同工型是1个基因的不同剪接方式造成的。
2.4 饲料脂肪水平对半滑舌鳎肝脏中ACC1基因表达的影响由图5可见,与对照组相比,3.5%鱼油组肝脏中ACC1 mRNA相对表达量显著降低(P<0.05);7.0%和10.0%鱼油组肝脏中ACC1 mRNA相对表达量进一步降低,显著低于对照组和3.5%组(P<0.05),同时7.0%和10.0%鱼油组间虽差异不显著(P>0.05),但在数值上10.0%鱼油组低于7.0%鱼油组。
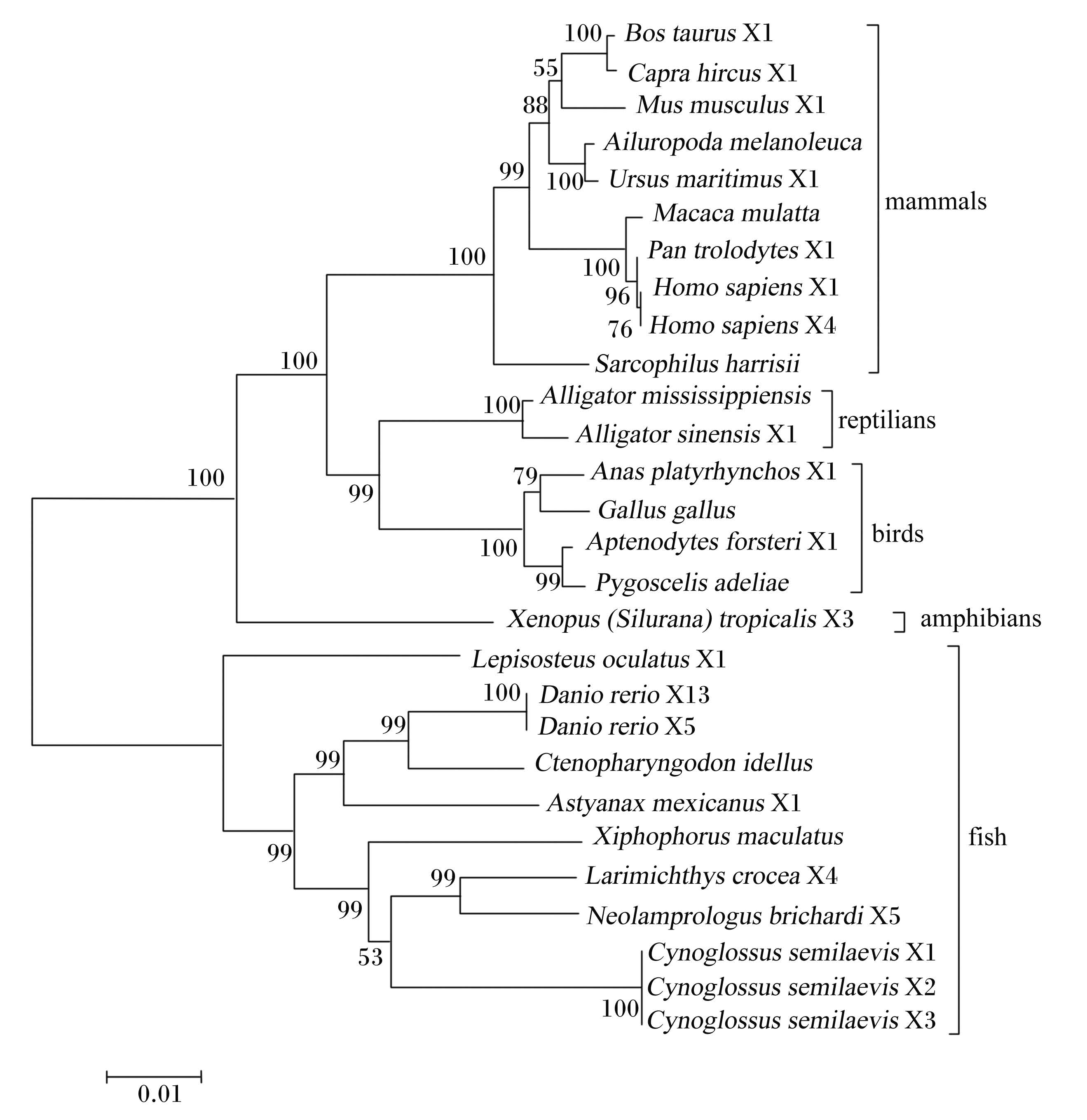 | 节点上的数字是自展值,物种拉丁文后接的"X+阿拉伯数字",表示该物种ACC1不同的同工型。The number on the node was the bootstrap value,and the"X+Arabic numeral"behind the Latin name mean the species'different isoforms.mam-mals:哺乳类;reptilians:爬行类;birds:鸟类;amphibians:两栖类;fish:鱼类;Bos taurus:牛;Capra hircus:山羊;Mus musculus:小鼠;Ailuropoda melanoleuca:大熊猫;Ursus maritimus:北极熊;Macaca mulatta:普通猕猴;Pan trolodytes:黑猩猩;Homo sa-piens:智人;Sarcophilus harrisii:袋獾;Alligator mississippiensis:美国短吻鳄;Alligator sinensis:扬子鳄;Anas platyrhynchos:绿头鸭;Gallus gallus:原鸡;Aptenodytes forsteri:帝企鹅;Pygoscelis adeliae:阿德利企鹅;Xenopus (Silurana) tropicalis:非洲爪蟾;Lepisosteus oculatus:眼斑雀鳝;Danio rerio:斑马鱼;Ctenopharyngodon idellus:草鱼;Astyanax mexicanus:墨西哥脂鲤;Xi-phophorus maculatus:月光鱼;Larimichthys crocea:大黄鱼;Neolamprologus brichardi:布氏新灿鲷。 图4 基于ACC1氨基酸序列的系统发育树 Fig. 4 Phylogenetic tree based on ACC1 amino acid sequences |
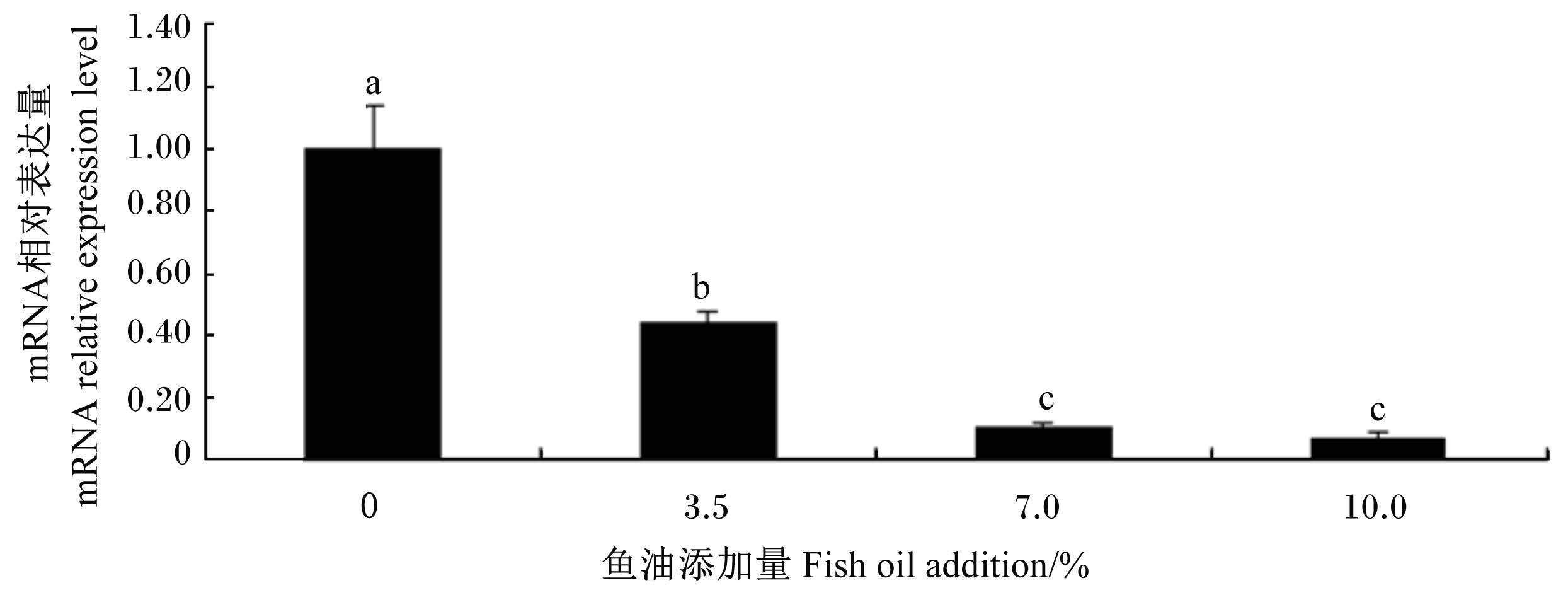 | 图5 饲料脂肪水平对半滑舌鳎ACC1基因表达的影响 Fig. 5 Effects of dietary fat level on ACC1 gene expression of half-smooth tongue sole (Cynoglossus semilaevis) |
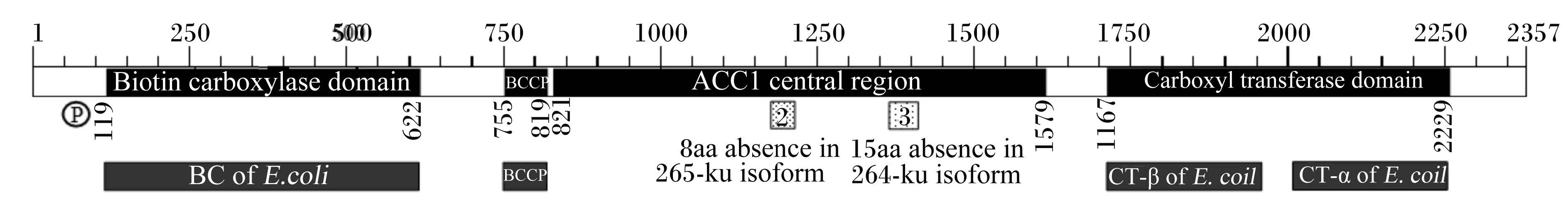
半滑舌鳎ACC1氨基酸序列与其他脊椎动物相比具有相似的结构特点,其主要结构域见图6。
由图6可见,半滑舌鳎ACC1由1个基因编码,具有3个结构域,分别对应原核生物的3个基因——BC、BCCP和CT,它们组成ACC1的3个亚基。BC域包含1个ATP结合位点,甘氨酸富集区(GGGGKG)被认为是人ACC1和ACC2的ATP结合域[6, 7],半滑舌鳎的ATP结合位点是Glu316~Glu321,与草鱼[12]和人类[6, 7]ATP结合域完全相同,说明这段序列高度保守。生物素酶的一个重要特征就是有1个生物素结合位点序列Met-Lys-Met的存在,这个区域有很保守的功能[3, 15, 16]。半滑舌鳎ACC1的BCCP域位于氨基酸序列的第755~819位点,唯一的4肽序列Val-Met-Lys-Met位于氨基酸残基的第785~788位点(图2)。 因此,脊椎动物的生物素结合位点高度保守[17]。人ACC1中区域SFSEIMQPWAQTVVVGRARLGGIPVGV和ACC2中区域SFKEIMAPWAQTVVTGRARLGGIPVGV被认为是辅酶A结合位点[6, 7],在半滑舌鳎CT域中有相似的序列1969SFMEIMKPWAQSVVVGRARLGGIPTGV1995。
 |
阴影框2和3表示可变剪接。Shaded boxes 2 and 3 showed alternative splicing.
Biotin carboxylase domain:生物素羧化酶结构域;ACC1 central region:ACC1中心区域;Carboxyl transferase domain:羧基转移酶结构域;BCCP:生物素羧基载体蛋白,biotin carboxyl carrier protein;BC of E.coil:大肠杆菌BC结构域;CT-β of E.coil:大肠杆菌CT-β结构域;CT-α of E.coil:大肠杆菌CT-α结构域;8aa absence in 265-ku isoform:缺少8个氨基酸分子质量为265 ku的同工型:15aa absence in 264-ku isoform:缺少15个氨基酸分子质量为264 ku的同工型。 图6 半滑舌鳎ACC1蛋白3个结构域与细菌ACC1蛋白的3个亚基比较 Fig. 6 Comparison of three domains of ACC1 for half-smooth tongue sole and three subunits of ACC1 for bacteria |
哺乳动物的ACC1基因共有54个编码蛋白质的外显子,在中心区域,编码8个氨基酸的外显子E28在大鼠[18]、小鼠[19]和绵羊[1]均存在可变剪接。本研究中,半滑舌鳎ACC1的中心区域位于氨基酸残基的第821~1 579位点(图6),该区域存在可变剪接,形成分子质量分别为266、265和264 ku的3个同工型,与分子质量为266 ku的ACC1相比,分子质量为265和264 ku的ACC1分别少8和15个氨基酸,这与在大鼠[18]、小鼠[19]和绵羊[1]中发现的情况类似。
3.2 半滑舌鳎ACC1基因的组织表达哺乳动物ACC1基因在各种组织中均表达,但在肝脏、脂肪、哺乳期乳腺等生脂组织中表达量最高[1]。半滑舌鳎ACC1在本试验检测的10种组织中均有表达,其中,肝脏和全脑等组织中表达量显著高于其他组织,与哺乳动物ACC1的组织表达规律相类似。
3.3 营养素对ACC1基因表达的调控对高等动物的研究表明,饲料营养素可通过调控相关基因表达控制动物体脂的沉积,高糖饲料能促进脂肪的合成,其作用涉及基因转录、mRNA加工和稳定多个层次[20];脂肪酸作为转录因子的配体参与了动物体内多种脂肪代谢相关基因表达的调节[21]。研究表明,脂肪酸对生脂基因表达有抑制作用,但这种作用与脂肪酸种类关系密切,饲料中添加多不饱和脂肪酸(PUFA)可降低鼠肝脏ACC等生脂基因mRNA的相对表达量[22]。饲料中脂肪种类和水平对鱼类生脂基因表达有显著影响,黑鲷(Sparus macrocephalus)饲料中当n-3 PUFA含量在0.92%以上时,脂肪酸合酶(FAS)基因表达量显著下降[23]。草鱼饲喂分别以猪油和鱼油为主要脂肪源的饲料后发现,富含n-3 PUFA的鱼油组肝胰脏FAS、ACC等生脂基因mRNA的相对表达量显著低于猪油组[24]。鲫鱼肝胰脏ACC1和FAS等生脂基因mRNA相对表达量随饲料脂肪水平的升高而降低[25]。本研究中,饲料中添加鱼油显著抑制了半滑舌鳎肝脏ACC1基因的表达。因此,当饲料中提供充足的PUFA时,半滑舌鳎可以优先利用,从而减少脂肪酸的从头合成,以节省能量。
4 结 论本研究从半滑舌鳎肝脏中克隆了ACC1基因全长cDNA,得出半滑舌鳎ACC1的主要功能位点为ATP结合位点、生物素结合位点、辅酶A结合位点,与其他脊椎动物相比基本保守。半滑舌鳎ACC1基因主要在肝脏和全脑等生脂组织中表达,饲料中添加鱼油显著抑制其肝脏ACC1基因的表达,且抑制作用与鱼油添加量呈正相关。
| [1] | BARBER M C,PRICE N T,TRAVERS M T.Structure and regulation of acetyl-CoA carboxylase genes of metazoa[J]. Biochimica et Biophysica Acta:Molecular and Cell Biology of Lipids,2005,1733(1):1-28. ( 4) 4)
|
| [2] | TONG L.Acetyl-coenzyme a carboxylase:crucial metabolic enzyme and attractive target for drug discovery[J]. Cellular and Molecular Life Sciences,2005,62(16):1784-1803. ( 1) 1)
|
| [3] | JITRAPAKDEE S,WALLACE J C.The biotin enzyme family:conserved structural motifs and domain rearrangements[J]. Current Protein & Peptide Science,2003,4(3):217-219. ( 2) 2)
|
| [4] | LÓPEZ-CASILLAS F,BAI D H,LUO X C,et al.Structure of the coding sequence and primary amino acid sequence of acetyl-coenzyme A carboxylase[J]. Proceedings of the National Academy of Sciences of the United States of America,1988,85(16):5784-5788. ( 2) 2)
|
| [5] | TAKAI T,YOKOYAMA C,WADA K,et al.Primary structure of chicken liver acetyl-CoA carboxylase deduced from cDNA sequence[J]. Journal of Biological Chemistry,1988,263(6):2651-2657. ( 2) 2)
|
| [6] | ABU-ELHEIGA L,ALMARZA-ORTEGA D B,BALDINI A,et al.Human acetyl-CoA carboxylase 2 Molecular cloning,characterization,chromosomal mapping,and evidence for two isoforms[J]. Journal of Biological Chemistry,1997,272(16):10669-10677. ( 5) 5)
|
| [7] | ABU-ELHEIGA L,JAYAKUMAR A,BALDINI A,et al.Human acetyl-CoA carboxylase:characterization,molecular cloning,and evidence for two isoforms[J]. Proceedings of the National Academy of Sciences of the United States of America,1995,92(9):4011-4015. ( 4) 4)
|
| [8] | ZHANG H L,YANG Z R,SHEN Y,et al.Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase[J]. Science,2003,299(5615):2064-2067. ( 1) 1)
|
| [9] | MAO J,MARCOS S,DAVIS S K,et al.Genomic distribution of three promoters of the bovine gene encoding acetyl-CoA carboxylase alpha and evidence that the nutritionally regulated promoter Ⅰ contains a repressive element different from that in rat[J]. Biochemical Journal,2001,358(Pt.1):127-135. ( 1) 1)
|
| [10] | BARBER M C,TARVERS M T.Cloning and characterisation of multiple acetyl-CoA carboxylase transcripts in ovine adipose tissue[J]. Gene,1995,154(2):271-275. ( 1) 1)
|
| [11] | BADAOUI B,SERRADILLA J M,TOMÀS A,et al.Goat acetyl-coenzyme a carboxylase α:molecular characterization,polymorphism,and association with milk traits[J]. Journal of Dairy Science,2007,90(2):1039-1043. ( 1) 1)
|
| [12] | CHENG H L,JI N J,PENG Y X,et al.Molecular characterization and tissue-specific expression of the acetyl-CoA carboxylase α gene from grass carp,Ctenopharyngodon idella[J]. Gene,2011,487(1):46-51. ( 2) 2)
|
| [13] | DIEFFENBACH C W,DVEKSLER G S.PCR primer:a laboratory manual[M]. Plainview NY:Cold Spring Harbor Laboratory Press,1995. ( 1) 1)
|
| [14] | TAMURA K,STECHER G,PETERSON D,et al.MEGA6:molecular evolutionary genetics analysis version 6.0[J]. Molecular Biology and Evolution,2013,30(12):2725-2729. ( 1) 1)
|
| [15] | JITRAPAKDEE S,ST MAURICE M,RAYMENT I,et al.Structure,mechanism and regulation of pyruvate carboxylase[J]. Biochemical Journal,2008,413(3):369-387. ( 1) 1)
|
| [16] | LEE C K,CHEONG H K,RYU K S,et al.Biotinoyl domain of human acetyl-CoA carboxylase:structural insights into the carboxyl transfer mechanism[J]. Proteins:Structure,Function,and Bioinformatics,2008,72(2):613-624. ( 1) 1)
|
| [17] | BAI D H,MOON T W,LOPEZ-CASILLAS F,et al.Analysis of the biotin-binding site on acetyl-CoA carboxylase from rat[J]. European Journal of Biochemistry,1989,182(2):239-245. ( 1) 1)
|
| [18] | BARBER M C,POOLEY L,TRAVERS M T.Developmental regulation of alternatively spliced acetyl-CoA carboxylase-alpha mRNAs encoding isozymes with or without an eight amino acid domain upstream of the Ser-1200 phosphorylation motif in the mammary gland[J]. Journal of Molecular Endocrinology,2001,27(3):349-356. ( 2) 2)
|
| [19] | SALLES J,SARGUEIL F,KNOLL-GELLIDA A,et al.Acetyl-CoA carboxylase and SREBP expression during peripheral nervous system myelination[J]. Biochimica et Biophysica Acta:Molecular and Cell Biology of Lipids,2003,1631(3):229-238. ( 2) 2)
|
| [20] | KOO H Y,WALLING M A,CHUNG B H,et al.Dietary fructose induces a wide range of genes with distinct shift in carbohydrate and lipid metabolism in fed and fasted rat liver[J]. Biochimica et Biophysica Acta:Molecular Basis of Disease,2008,1782(5):341-348. ( 1) 1)
|
| [21] | TAI C C,DING S T.N-3 polyunsaturated fatty acids regulate lipid metabolism through several inflammation mediators:mechanisms and implications for obesity prevention[J]. Journal of Nutritional Biochemistry,2010,21(5):357-363. ( 1) 1)
|
| [22] | KAJIKAWA S,HARADA T,KAWASHIMA A,et al.Highly purified eicosapentaenoic acid prevents the progression of hepatic steatosis by repressing monounsaturated fatty acid synthesis in high-fat/high-sucrose diet-fed mice[J]. Prostaglandins,Leukotrienes and Essential Fatty Acids,2009,80(4):229-238. ( 1) 1)
|
| [23] | 马晶晶,邵庆均,许梓荣,等.n-3高不饱和脂肪酸对黑鲷幼鱼生长及脂肪代谢的影响[J]. 水产学报,2009,33(4):639-649. ( 1) 1)
|
| [24] | 李超.n-3高不饱和脂肪酸对草鱼生长、脂代谢及健康状况的影响[D]. 硕士学位论文.杨凌:西北农林科技大学,2013. ( 1) 1)
|
| [25] | ZHOU J C,HAN D,JIN J Y,et al.Compared to fish oil alone,a corn and fish oil mixture decreases the lipid requirement of a freshwater fish species,Carassius auratus gibelio[J]. Aquaculture,2014,428/429:272-279. ( 1) 1)
|




