2. 中国科学院亚热带农业生态研究所, 亚热带农业生态过程重点实验室, 长沙 410125
2. Key Laboratory of Subtropical Agro-Ecological Processes, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha 410125, China
印记基因通常是指仅一方亲本来源的同源基因表达,而来自另一亲本的不表达,因而导致后代体细胞中2个亲本来源的等位基因有不同的表达方式。自印记基因被发现以来,研究人员对哺乳动物印记基因的个数、印记基因的调控机制及印记基因对机体不同组织器官的调节作用已进行了一定的研究。胎盘中的印记基因表达受胎儿性别、妊娠期、年龄、分娩方式等多种因素影响[1]。作为生命的开始,胎盘和胚胎的生长发育状态与哺乳动物出生后的健康状态有着不可忽视的联系,因此探索印记基因对胎盘和胚胎发育的调控机制可为哺乳动物后期的生长发育调控与疾病预防等提供科学依据。鉴于此,本文基于文献报道综述了印记基因的主要特点,以及近年来印记基因对哺乳动物胚胎和胎盘发育的研究进展,旨在为系统深入开展印记基因对胎盘和胚胎发育的调控机制研究奠定基础。
1 基因组印记与印记基因1960年Crouse[2]在研究尖眼蕈蚊(Sciara)时发现其2条X染色体中只有来自母系的等位基因表达活性,而来自父系的等位基因始终处于沉默状态,进而第一次提出了基因组印记的概念。当时“基因组印记”是用来形容在节肢动物物种的性别决定中起作用的父源特定染色体缺失[3]。随着科学研究的深入,如今基因组印记用来形容由表观遗传修饰决定,来源于双亲的基因所呈现的特异性表达,存在基因组印记的基因也就被称之为印记基因。正常情况下印记基因只表达一方亲本来源的等位基因,而另一亲本被一系列表观遗传修饰后沉默。基因组印记区别于与自身性别相关的伴性遗传,属于非孟德尔遗传学的表观遗传学领域。基因组印记体系中把父系等位基因抑制而母系等位基因表达的基因定义为父系印记基因,父系等位基因表达而母系等位基因抑制的基因定义为母系印记基因。基因组印记的遗传理论认为母系印记基因促进胎儿和胎盘增长,父系印记基因则限制胎体生长[4]。
关于哺乳动物基因组印记最早可以追溯到1983年McGrath等[5]在利用核移植技术培育小鼠时,发现孤雌胚胎中2套基因组均为同一亲本,一亲本基因双倍表达,而另一亲本基因缺失,组合胚可以短期发育,但最终会死亡,由此推测母源基因组和父源基因组都是子代生长必需的。之后,哺乳动物基因胰岛素样生长因子2(insulin-like growth factor 2,IGF2)[6-9]、胰岛素样生长因子2受体(insulin-like growth factor-2 receptor,IGF2R)[5]和H19[9]也在小鼠上被发现。继而在2015年11月,人和小鼠中包括蛋白质编码基因、非编码RNA转录本以及小核仁RNA(small nucleolar RNA,snoRNA)和小RNA(micro RNA,miRNA)在内的已被确定的印记基因分别已达100和151个,且数量还在不断上升(http://www.mousebook.org/imprinting-gene-list,http://igc.otago.ac.nz/home.html)。随后,随着研究范围的进一步拓宽,研究人员相继从猪、牛和绵羊等动物中发现了印记基因的存在,同时也有研究指出卵生哺乳动物可能缺乏印记基因,如鸭嘴兽和针鼹[10]。
2 印记基因表达的主要特点印记基因的表达虽然与经典的孟德尔遗传规律并不完全吻合,但却有别于普通基因的表达特征。
2.1 以基因簇方式出现印记基因簇通常包含多个印记基因和至少1个非编码RNA(noncoding RNA,ncRNA)印记。所有的簇内都有1个印记控制中心(imprinted control center,ICR),ICR通常是1~5 kb,来源于双亲等位基因中的1个ICR会被DNA甲基化标记,这些差异甲基化区域(differentially methylated region,DMR)使双亲等位基因出现差异性表达,从而在整个印记簇内调节印记。其次,印记簇内至少有1个非编码RNA由母系等位基因和多个父系蛋白质编码基因表达。最后,印记基因簇可能通过以下4种机制进行调节:通过CpG岛(CpG island,CpGs)或启动子的差异甲基化,关闭染色体构象形成异染色质;差异性的将沉默因子结合到顺式沉默元件上,沉默因子与未被甲基化的顺式沉默元件结合则抑制基因的表达;通过CCCTC结合因子(CCCTC-binding factor,CTCF)结合绝缘子以阻断共享的增强子元件[11];反义转录本与CpGs或启动子的甲基化联合作用机制调控正义基因的表达。
2.2 表达时间具有特异性普通基因的表达水平通常与细胞周期中的复制时间相关,但印记基因并不遵循这一规律,印记亲源等位基因的复制具有不同步性的鲜明特征。
2.3 表达空间具有特异性印记基因在有的组织中印记表达,但在其他组织上又呈现非印记特点。有研究发现,父系印记基因溶质载体家族蛋白22成员3(solute carrier family 22 member 3,Slc22a3)在小鼠胚胎发育早期在胎盘中呈现特异性印记;母系印记基因溶质载体家族蛋白38成员4(solute carrier family 38 member 4,Slc38a4)则在小鼠除肝脏和肠以外的所有组织中表达[12]。父系印记基因生长因子受体结合蛋白10(growth factor receptor-bound protein 10,Grb10)在小鼠大脑中表达,而母系等位基因则在几乎所有的组织和器官表达[13]。
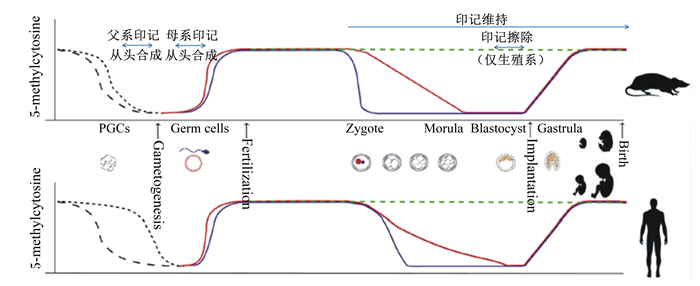
3 印记基因的擦除、重建与表观遗传修饰印记是在子代雌雄配子发生过程中建立的,是一个DNA甲基化的动态变化过程,包含印记在性腺中的擦除、印记的重建和印记重建后的维持3个部分(图 1)[14]。雌雄配子的甲基化程度差异显著,卵子的甲基化程度低,精子的甲基化程度较高,但均低于体细胞,卵子和精子的甲基化程度的差异被认为可能是配子产生印记的机制[15]。
|
5-methylcytosine:5-甲基胞嘧啶;PGCs:原始生殖细胞;Gametogenesis:配子发生;Germ cells:生殖细胞;Fertilization:受精;Zygote:受精卵;Morula:桑葚胚;Blastocyst:囊胚;Implantation:定植;Gastrula:原肠胚;Birth:出生。 图1 胚胎发育各阶段细胞的甲基化程度及其印记状态 Fig. 1 The methylation degree of cells in different stages of embryonic development and its imprinting status |
配子从亲本携带的第一代印记在受精、卵裂期间会一直保持,只有到8细胞期至囊胚期才会通过大规模去甲基化在性腺中被擦除。
对于印记的重建(获得第2代印记),现有观点认为父系印记的重建发生在精子发生前,母系印记发生在卵子发生后[16]。研究者们在小鼠中发现,去甲基化过程在小鼠妊娠E12~13期间完成。原始生殖细胞(prmordial germ cells,PGCs)在交配的7 d之后从外胚层迁移到性腺,10.5 d时在性腺发育成配子,13.5 d时雌性生殖细胞进入减数分裂,而雄性生殖细胞的有丝分裂则被抑制。在这一过程中,生殖细胞进行了一系列重要的表观遗传改写。交配后第8天,机体通过减少DNA的甲基化修饰和组蛋白修饰促进CpGs的迁移(可能是被动的过程)[17]。大约在10.5 d生殖细胞迁徙至性腺后,CpGs发生主动且快速的去甲基化作用,但ICR还会持续大约1 d的甲基化标记。有研究表明,活化诱导胞嘧啶核苷脱氨酶(activation-induced cytidine deaminase,AID)有助于第2次甲基化,并认为组蛋白置换在这个活动过程中也扮演了重要的角色[17]。亲本携带的印记在去甲基化的 过程中都会被擦除,这说明亲代的遗传印记对子代并没有太大影响。
4 印记基因对胚胎和胎盘发育的调控研究表明,印记基因既可调控母体通过胎盘供给子代的营养物质[4, 18],也可调控小鼠胚胎的生长发育(基因靶向试验)[19],且印记基因参与胚胎细胞的各种生物学过程[20-21]。例如印记基因溶质载体家族蛋白22成员2(solute carrier family 22 member 2,Slc22a2)可编码有机阳离子的转运蛋白[22],电压门控钾通道亚家族(Kcn)和Kcne2可编码钾离子的转运蛋白[23],且特定的钾离子通道阻滞剂会抑制人合体滋养细胞中绒毛膜促性腺激素的分泌[24],进而可调控母体营养物质和代谢前体物的转运。印记基因溶质载体家族蛋白38(solute carrier family 38,Slc38)转运家族对胎盘中钠离子依赖性氨基酸转运系统起主要的调节作用,其中Slc38a4编码中性与碱性氨基酸转运蛋白,其与亚型蛋白钠耦合中性氨基酸转运蛋白4(Snat4)组成的异构体Slc38a4/Snat4已证实存在于人的胎盘中[25],在大鼠和小鼠的胎盘中也检测到该异构体的表达[26-28],由此提示印记基因可以通过控制胎盘中氨基酸的转运控制母体与子代之间的营养物质传输。印记基因细胞周期蛋白依赖性激酶抑制剂1c (Cdkn1c)可编码细胞周期抑制剂,而细胞周期抑制剂的表达与血管内皮生长因子的表达呈负相关[29],因此Cdkn1c可通过调控血管内皮生长因子在供血丰富的胚胎组织及增殖期子宫内膜的血管生成中发挥作用。并且,研究发现印记基因Grb10在介导胰岛素、胰岛素样生长因子调控细胞增殖和凋亡中起着重要的作用[30-31]。此外,有文献报道IGF2是心室心肌细胞增殖的主要有丝分裂信号[32]。研究还发现,印记基因甘氨酸脒基转移酶编码基因(glycine amidinotransferase,Gatm)可调控肌酸的合成[33],同时参与胚胎组织器官的形成。此外,至少有25%印记基因编码反义RNA[如IGF2R编码IGF2R反义序列RNA(antisense of IGF2R RNA,Air)]、snoRNA[如SNRPN编码HBⅡ-52和HBⅡ-85)和miRNA[如转座子1(retrotransposon-like 1,Rtl1)编码miRNA-127和miRNA-136][31]。
目前对印记基因印记机制的理解主要来自于6个印记区,包括4个母系印记区[IGF2R/Air、印记中心2(IC2)/Kcnq1、G蛋白α亚单位(Gnas)及Prader-Willi综合征印记区(PWS/AS)和2个父系印记区[IGF2/H19和Dlkl1(delta-like 1 homologue)][34]。其中调控胚胎和胎盘发育的主要是H19/IGF2印记区、Dlk1-3型去碘酶(type 3 deiodinase,Dio3)印记区和印记基因父系表达基因10(paternally expressed gene 10,Peg10)。
4.1 H19/IGF2印记区H19和IGF2基因的表达相互耦合协同形成一个相互印记区(IGF2/H19),位于人类11号染色体[35]和小鼠的7号染色体。H19/IGF2印记区被认为参与胎盘的形成和胚胎的发育。印记会限制H19的表达仅限于母源等位基因,而IGF2仅转录父系等位基因[36]。
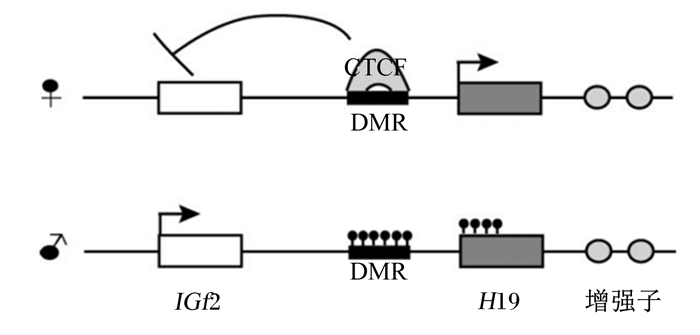
|
图2 H19/IGF2印记机制模型 Fig. 2 H19/IGF2imprinting mechanism model[38] |
H19/IGF2印记机制的模型(图 2)由1个ICR、两侧的基因、位于下游的增强子、CTCF和1个黏性复合体操控的远端染色体交互作用构成[37-38]。近期已有试验证明这种发挥交互作用的复合体是桩蛋白,它可以调节H19和IGF2之间的远程互作[39]。CTCF、母系未甲基化的ICR和block(多序列无空对比而产生的蛋白质序列)相互结合,通过增强子到IGF2的启动子位置发挥作用[40-41]。而H19的ICR父系甲基化抑制其与CTCF的结合,使得增强子激活父系染色体IGF2基因启动子[42-43]。保持这种印记模式是胚胎细胞生长和发育的关键[44]。
H19/IGF2印记区ICR的缺失会导致中胚层的组织特异性丢失[45],因此该印记区被认为与胚胎形成相关。Angiolini等[46]研究发现H19的mRNA能使许多与细胞迁移、血管形成和胎盘血流有关的基因上调,敲除H19会导致胎盘各层组织异常增生,因此认为H19在改变胎盘功能和胚胎生长、发育及个体行为发展方面起重要作用。IGF2基因的转录本在胚胎迷路滋养层中特异性表达[47],主要作用是调控胎盘、胚胎之间的营养供需平衡[48],参与氨基酸主动转运系统的代偿[49]。IGF2的启动子P0控制小鼠迷路滋养层细胞,启动子P0的缺失会降低胎盘部位IGF2 mRNA的表达,降低胎盘的被动扩散功能[49],且由IGF2父系基因调节的血清浓度和mRNA的表达量与婴儿出生重呈正相关[50-51]。胎儿生长受限还与5-甲基胞嘧啶胎盘和脐带IGF2基因的DMR处的作用有关[50, 52-54],因此认为IGF2参与了胚胎血流和营养的供给。
4.2 Dlk1-Dio3印记区Dlk1-Dio3印记区位于人14号染色体、小鼠12号染色体、绵羊18号染色体,并且在这3种哺乳动物中高度保守。该印记区包含3个父系表达蛋白质编码基因Dlk1、父系表达基因11(paternally expressed gene 11,Peg11)和Dio3以及一些母系表达非编码转录本,如母系表达基因3(maternally expressed gene 3,Meg3)/Gtl2(gene trap locus 2)、miRNA和C/D snoRNA[55]。
20世纪80年代有学者发现经杂交试验获得的单亲二倍体小鼠均在围产期死亡,并伴随胎盘、软骨、成骨组织以及骨骼肌等多种器官的生长发育缺陷[55],由此提示Dlk1-Dio3印记区对于小鼠胚胎和胎盘的生长发育调控至关重要。Peg11主要表达于胚胎和胎盘组织[56],在真哺乳亚纲动物尿囊绒毛膜中参与胎盘与胚胎的信息传递[57]。Dio3可编码Ⅲ型脱碘酶降解甲状腺激素,有研究发现Ⅲ型脱碘酶在健康哺乳动物的胎盘和子宫组织中高度表达,其原因可能是为了保护胚胎组织免受过高浓度甲状腺激素带来的伤害[58],此外,缺乏Dio3基因的新生小鼠发育后期中枢甲状腺机能减退,甚至产生甲状腺毒症[59]。Gtl2在小鼠E3.5期就出现表达且为印记表达[60],当基因敲除父本染色体Glt2的DMR到第5外显子共10 kb的区域后,小鼠胚胎出现了严重的生长迟缓并伴随围产期较高死亡率[61],当基因敲除母本染色体Glt2第1外显子到第5外显子共5 kb的区域后,下游的长链非编码RNA表达完全受到抑制,并导致小鼠围产期死亡[62],由此提示Dlk1-Dio3印记区内的非编码RNA转录本参与调控胎盘和胚胎生长。基因敲除Meg3后,小鼠胚胎脑中血管内皮生长因子及其Ⅰ型受体的表达显著提高[63],脑血管增加[64],表明Meg3参与神经与代谢调控,可能在抑制肿瘤发生方面也发挥了一定的作用[65]。
4.3 Peg10Peg10位于人类7号染色体、小鼠6号染色体和牛4号染色体上。Peg10的表达主要分布于胎盘、卵巢、睾丸及心、肺、脑等组织中,在迷路滋养细胞形成、胚胎发育、妊娠期新陈代谢和正常妊娠免疫耐受的建立等方面也发挥着重要作用。Peg10配子DMR DNA甲基化的缺失会降低胚胎的发育能力、减少迷路滋养层的体积[66]。有报道指出Peg10异常甲基化可使克隆牛胎盘异常发育,增加早期流产风险[67]。敲除Peg10基因可导致小鼠胎盘发育缺陷[47]、胚胎发育异常、胚胎中成胶质细胞及迷路层缺失,甚至早期胚胎死亡[68]。Peg10维持在一定水平可以促进孕激素及绒毛膜促性腺激素的合成,有利于胎盘定植及胚胎发育。Peg10在妊娠早期和中期表达量上升与早期蜕膜与绒毛滋养细胞分化、发育及胎盘形成有关,Peg10的ICR被认为是妊娠中期胎盘功能的关键调节因子[66]。Peg10妊娠晚期表达量下降[69]与胎盘钙化、葡萄糖转运等有密切关联[70]。妊娠晚期胎盘组织中Peg10异常表达不仅降低胎盘效率、胎儿和胎盘质量比[71],还会增加妊娠期患子痫前期和胎儿宫内发育迟缓的风险[72]。
4.4 其他印记基因对胚胎和胎盘发育的调控父系表达基因1(paternally expressed gene1,Peg1)位于人7号染色体,在所有胎儿组织表达[73],有试验显示Peg1在胎盘组织上的表达并不依赖于启动子区甲基化水平的变化,而可能是Peg1基因2个启动子之间的父系表达非编码RNA父系表达基因转录本1(Peg1-As)在生长发育过程中参与调控Peg1的表达[74]。父系表达基因3(paternally expressed gene3,Peg3)定位于小鼠7号染色体,敲除Peg3后,小鼠胎盘体积缩小[49],有报道指出Peg3突变的小鼠,其胎盘较小,胚胎出生后体重较低[75],另有试验发现Peg3在人胎盘中的高表达受其启动子区甲基化的影响,与婴儿的初生重存在正相关[76]。L3mbtl1是位于人20号染色体上的父系表达基因,在生殖细胞和生殖干细胞早期阶段均有表达,L3mbtl1缺陷可抑制生殖干细胞染色质转录,并影响胚胎干细胞向滋养外胚层分化。在妊娠早期胎盘组织中L3mbtl1表达量下调,可能会增加流产、胚胎停育的风险[67]。Mash2基因可以调节成浆细胞的发育,这对胎盘的生长发育起重要作用[36]。Kcnq1基因簇中的基因只在胎盘中表达印记,维持由DNA甲基化转移酶1(Dnmt1)修饰突变的胚胎中印记的表达[55],Kcnq1配子DMR的丢失与滋养层巨细胞的膨胀密切相关[61]。Phlda2基因的表达在宫内发育迟缓婴儿的胎盘中上调[77-78],提示该基因可作为一种负增长调节剂。在小鼠早期胚胎中能发育相关基因3(Dppa3)可保护5-甲基胞嘧啶不被TET(ten-eleven translocation)蛋白催化氧化[79],提示Dppa3可通过调控DNA甲基化状态调控胚胎发育中的印记状态[80]。
5 小 结综上所述,哺乳动物胎盘和胚胎的生长发育受多种印记基因调控,其功能异常可导致胚胎的发育受限。印记基因在胎盘和胚胎的表达模式与成年动物组织中的表达模式存在显著差异,由此表明哺乳动物胎盘及胚胎中印记基因具有至关重要的调控功能。但目前研究多侧重于人和小鼠,大部分研究成果也是基于人和小鼠这2种模型,加强对农业经济型动物(猪牛羊等)胚胎和胎盘印记基因的研究力度将有助于改善其繁殖性能,减少流产损失和幼畜出生缺陷。另外,除了对H19、IGF2、IGF2R等早期发现的印记基因的序列、ICR、DNA甲基化状态研究的较为透彻外,其余大部分印记基因的印记机制尚不明确。研究人员对于印记基因与胎盘、胚胎之间的互作研究也很薄弱。针对现有研究内容的盲区,可以预见不同哺乳动物印记基因的完全确定、不同印记簇或印记之间的机制与互作关系、不同印记对生殖器官发育的调控作用及其作用机制等方面的研究将成为相关研究领域的热点。
| [1] |
BURTON G J, SEBIRE N J, MYATT L, et al. Optimising sample collection for placental research[J].
Placenta, 2014, 35(1): 9–22.
( 0) 0)
|
| [2] |
CROUSE H V. The controlling element in sex chromosome behavior in Sciara[J].
Genetics, 1960, 45(10): 1429–1443.
( 0) 0)
|
| [3] |
CROUSE H V, BROWN A, MUMFORD B C. L-Chromosome inheritance and the problem of chromosome "imprinting" in Sciara (Sciaridae,Diptera)[J].
Chromosoma, 1971, 34(3): 324–339.
( 0) 0)
|
| [4] |
MOORE T, HAIG D. Genomic imprinting in mammalian development:a parental tug-of-war[J].
Trends in Genetics, 1991, 7(2): 45–49.
( 0) 0)
|
| [5] |
MCGRATH J, SOLTER D. Nuclear transplantation in the mouse embryo by microsurgery and cell fusion[J].
Science, 1983, 220(4603): 1300–1302.
( 0) 0)
|
| [6] |
BARLOW D P, STÖGER R, HERRMANN B G, et al. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus[J].
Nature, 1991, 349(6304): 84–87.
( 0) 0)
|
| [7] |
DECHIARA T M, ROBERTSON E J, EFSTRATIADIS A. Parental imprinting of the mouse insulin-like growth factor Ⅱ gene[J].
Cell, 1991, 64(4): 849–859.
( 0) 0)
|
| [8] |
FERGUSON-SMITH A C, CATTANACH B M, BARTON S C, et al. Embryological and molecular investigations of parental imprinting on mouse chromosome 7[J].
Nature, 1991, 351(6328): 667–670.
( 0) 0)
|
| [9] |
BARTOLOMEI M S, ZEMEL S, TILGHMAN S M. Parental imprinting of the mouse H19 gene[J].
Nature, 1991, 351(6322): 153–155.
( 0) 0)
|
| [10] |
RENFREE M B, HORE T A, SHAW G, et al. Evolution of genomic imprinting:insights from marsupials and monotremes[J].
Annual Review of Genomics and Human Genetics, 2009, 10: 241–262.
( 0) 0)
|
| [11] |
WAN L B, BARTOLOMEI M S. Regulation of imprinting in clusters:noncoding RNAs versus insulators[J].
Advances in Genetics, 2008, 61: 207–223.
( 0) 0)
|
| [12] |
周泉勇.大白猪和二花脸猪妊娠后期胎盘转录谱比较及印记基因鉴定研究[D].博士学位论文.武汉:华中农业大学,2009.
( 0) 0)
|
| [13] |
HIKICHI T, KOHDA T, KANEKO-ISHINO T, et al. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes;roles of brain-specific promoters and mouse-specific CTCF-binding sites[J].
Nucleic Acids Research, 2003, 31(5): 1398–1406.
( 0) 0)
|
| [14] |
MONK D. Germline-derived DNA methylation and early embryo epigenetic reprogramming:the selected survival of imprints[J].
The International Journal of Biochemistry & Cell Biology, 2015, 67: 128–138.
( 0) 0)
|
| [15] |
白莉雅, 肖遥, 滑国华, 等. 表观遗传学在动物繁育上的应用研究[J].
中国奶牛, 2010(9): 19–23.
( 0) 0)
|
| [16] |
BARLOW D P, BARTOLOMEI M S. Genomic imprinting in mammals[J].
Cold Spring Harbor Perspectives in Biology, 2014, 6(2): 1–20.
( 0) 0)
|
| [17] |
HAJKOVA P, ANCELIN K, WALDMANN T, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line[J].
Nature, 2008, 452(7189): 877–881.
( 0) 0)
|
| [18] |
CONSTÂNCIA M, KELSEY G, REIK W. Fertility resourceful imprinting[J].
Nature, 2004, 432(7013): 53–57.
( 0) 0)
|
| [19] |
MORISON I M, RAMSAY J P, SPENCER H G. A census of mammalian imprinting[J].
Trends in Genetics, 2005, 21(8): 457–465.
( 0) 0)
|
| [20] |
CHARALAMBOUS M, DA ROCHA S T, FERGUSON-SMITH A C. Genomic imprinting,growth control and the allocation of nutritional resources:consequences for postnatal life[J].
Current Opinion in Endocrinology,Diabetes,and Obesity, 2007, 14(1): 3–12.
( 0) 0)
|
| [21] |
WILKINSON L S, DAVIES W, ISLES A R. Genomic imprinting effects on brain development and function[J].
Nature Reviews Neuroscience, 2007, 8(11): 832–843.
( 0) 0)
|
| [22] |
KOEPSELL H. The SLC22 family with transporters of organic cations,anions and zwitterions[J].
Molecular Aspects of Medicine, 2013, 34(2/3): 413–435.
( 0) 0)
|
| [23] |
ABBOTT G W, TAI K K, NEVERISKY D, et al. KCNQ1,KCNE2,and Na+-coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability[J].
Science Signaling, 2014, 7(315): 22.
( 0) 0)
|
| [24] |
WILLIAMS J L R,FYFE G K,SIBLEY C P,et al.TEA-sensitive K+ channels regulate hCG secretion and production by human villous cytotrophoblast cells in vitro[C]//Pediatric research.Baltimore:International Pediatric Research Foundation,2007,62(3):387-387.
( 0) 0)
|
| [25] |
DESFORGES M, LACEY H A, GLAZIER S L, et al. SNAT4 isoform of system A amino acid transporter is expressed in human placenta[J].
American Journal of Physiology:Cell Physiology, 2006, 290(1): C305–C312.
( 0) 0)
|
| [26] |
NOVAK D, LEHMAN M, BERNSTEIN M, et al. SNAT expression in rat placenta[J].
Placenta, 2006, 27(4/5): 510–516.
( 0) 0)
|
| [27] |
SMITH R J, DEAN W, KONFORTOVA G, et al. Identification of novel imprinted genes in a genome-wide screen for maternal methylation[J].
Genome Research, 2003, 13(4): 558–569.
( 0) 0)
|
| [28] |
DESFORGES M, SIBLEY C P. Placental nutrient supply and fetal growth[J].
International Journal of Developmental Biology, 2010, 54(2/3): 377–390.
( 0) 0)
|
| [29] |
CESARIO J M, MALT A L, DEACON L J, et al. Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene,p57Kip2[J].
Human Molecular Genetics, 2015, 24(17): 5024–5039.
( 0) 0)
|
| [30] |
SHEN T L, GUAN J L. Grb7 in intracellular signaling and its role in cell regulation[J].
Frontiers in Bioscience, 2004, 9: 192–200.
( 0) 0)
|
| [31] |
MARCINIAK M. Imprinting genomowy u ssakow:najnowsze doniesienia[J].
Postapy Biologii Komórki, 2008, 35(2): 243–257.
( 0) 0)
|
| [32] |
SHEN H, CAVALLERO S, ESTRADA K D, et al. Extracardiac control of embryonic cardiomyocyte proliferation and ventricular wall expansion[J].
Cardiovascular Research, 2015, 105(3): 271–278.
( 0) 0)
|
| [33] |
STOCKLER-IPSIROGLU S, VAN KARNEBEEK C D M. Cerebral creatine deficiencies:a group of treatable intellectual developmental disorders[J].
Seminars in Neurology, 2014, 34(3): 350–356.
( 0) 0)
|
| [34] |
郭玲.人异常胎盘转录组分析及MAGEL2基因在猪胚胎中的印记研究[D].博士学位论文.武汉:华中农业大学,2013.
( 0) 0)
|
| [35] |
REIK W, DEAN W, WALTER J. Epigenetic reprogramming in mammalian development[J].
Science, 2001, 293(5532): 1089–1093.
( 0) 0)
|
| [36] |
BARTOLOMEI M S, FERGUSON-SMITH A C. Mammalian genomic imprinting[J].
Cold Spring Harbor Perspectives in Biology, 2011, 3(7): 322–330.
( 0) 0)
|
| [37] |
MACDONALD W A. Epigenetic mechanisms of genomic imprinting:common themes in the regulation of imprinted regions in mammals,plants,and insects[J].
Genetics Research International, 2012, 2012: 585024.
( 0) 0)
|
| [38] |
VERONA R I, MANN M R W, BARTOLOMEI M S. Genomic imprinting:intricacies of epigenetic regulation in clusters[J].
Annual Review of Cell and Developmental Biology, 2003, 19: 237–259.
( 0) 0)
|
| [39] |
MARÁŠEK P, DZIJAK R, STUDENYAK I, et al. Paxillin-dependent regulation of IGF2 and H19 gene cluster expression[J].
Journal of Cell Science, 2015, 128(16): 3106–3116.
( 0) 0)
|
| [40] |
BELL A C, FELSENFELD G. Methylation of a CTCF-dependent boundary controls imprinted expression of the IGF2 gene[J].
Nature, 2000, 405(6785): 482–485.
( 0) 0)
|
| [41] |
HARK A T, SCHOENHERR C J, KATZ D J, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/IGF2 locus[J].
Nature, 2000, 405(6785): 486–489.
( 0) 0)
|
| [42] |
MURRELL A, HEESON S, REIK W. Interaction between differentially methylated regions partitions the imprinted genes IGF2 and H19 into parent-specific chromatin loops[J].
Nature Genetics, 2004, 36(8): 889–893.
( 0) 0)
|
| [43] |
KURUKUTI S, TIWARI V K, TAVOOSIDANA G, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2[J].
Proceedings of The National Academy of Sciences of the United States, 2006, 103(28): 10684–10689.
( 0) 0)
|
| [44] |
ISHIDA M, MOORE G E. The role of imprinted genes in humans[J].
Molecular Aspects of Medicine, 2013, 34(4): 826–840.
( 0) 0)
|
| [45] |
IDERAABDULLAH F Y, THORVALDSEN J L, MYERS J A, et al. Tissue-specific insulator function at H19/IGF2 revealed by deletions at the imprinting control region[J].
Human Molecular Genetics, 2014, 23(23): 6246–6259.
( 0) 0)
|
| [46] |
ANGIOLINI E, FOWDEN A, COAN P, et al. Regulation of placental efficiency for nutrient transport by imprinted genes[J].
Placenta, 2006, 27(Suppl.): 98–102.
( 0) 0)
|
| [47] |
CONSTÂNCIA M, DEAN W, LOPES S, et al. Deletion of a silencer element in IGF2 results in loss of imprinting independent of H19[J].
Nature Genetics, 2000, 26(2): 203–206.
( 0) 0)
|
| [48] |
CONSTÂNCIA M, HEMBERGER M, HUGHES J, et al. Placental-specific IGF-Ⅱ is a major modulator of placental and fetal growth[J].
Nature, 2002, 417(6892): 945–948.
( 0) 0)
|
| [49] |
王增艳.玻璃化冷冻小鼠胚胎对印记基因H19/IGF2印记控制区甲基化状态及基因表达量的影响[D].博士学位论文.北京:北京协和医学院,2010.
( 0) 0)
|
| [50] |
ST-PIERRE J, HIVERT M F, PERRON P, et al. IGF2 DNA methylation is a modulator of newborn's fetal growth and development[J].
Epigenetics, 2012, 7(10): 1125–1132.
( 0) 0)
|
| [51] |
HOYO C, FORTNER K, MURTHA A P, et al. Association of cord blood methylation fractions at imprinted insulin-like growth factor 2(IGF2),plasma IGF2,and birth weight[J].
Cancer Causes & Control, 2012, 23(4): 635–645.
( 0) 0)
|
| [52] |
KOUKOURA O, SIFAKIS S, SOUFLA G, et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction[J].
International Journal of Molecular Medicine, 2011, 28(4): 481–487.
( 0) 0)
|
| [53] |
KOUKOURA O, SIFAKIS S, ZARAVINOS A, et al. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction[J].
Placenta, 2010, 32(1): 51–57.
( 0) 0)
|
| [54] |
BOURQUE D K, AVILA L, PEÑAHERRERA M, et al. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia[J].
Placenta, 2010, 31(3): 197–202.
( 0) 0)
|
| [55] |
DA ROCHA S T, EDWARDS C A, ITO M, et al. Genomic imprinting at the mammalian Dlk1-Dio3 domain[J].
Trends Genetics, 2008, 24(6): 306–316.
( 0) 0)
|
| [56] |
BRANDT J, VEITH A M, VOLFF J N. A family of neofunctionalized Ty3/gypsy retrotransposon genes in mammalian genomes[J].
Cytogenetic and Genome Research, 2005, 110(1/2/3/4): 307–317.
( 0) 0)
|
| [57] |
SEKITA Y, WAGATSUMA H, NAKAMURA K, et al. Role of retrotransposon-derived imprinted gene,Rtl1,in the feto-maternal interface of mouse placenta[J].
Nature Genetics, 2008, 40(2): 243–248.
( 0) 0)
|
| [58] |
HERNANDEZ A, MARTINEZ M E, FIERING S, et al. Type 3 deiodinase is critical for the maturation and function of the thyroid axis[J].
The Journal of Clinical Investigation, 2006, 116(2): 476–484.
( 0) 0)
|
| [59] |
GALTON V A, MARTINEZ E, HERNANDEZ A, et al. Pregnant rat uterus expresses high levels of the type 3 iodothyronine deiodinase[J].
The Journal of Clinical Investigation, 1999, 103(7): 979–987.
( 0) 0)
|
| [60] |
NOWAK K, STEIN G, POWELL E, et al. Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus[J].
Epigenetics, 2011, 6(8): 1012–1020.
( 0) 0)
|
| [61] |
TAKAHASHI N, OKAMOTO A, KOBAYASHI R, et al. Deletion of Gtl2,imprinted non-coding RNA,with its differentially methylated region induces lethal parent-origin-dependent defects in mice[J].
Human Molecular Genetics, 2009, 18(10): 1879–1888.
( 0) 0)
|
| [62] |
ZHOU Y L, CHEUNSUCHON P, NAKAYAMA Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene[J].
Development, 2010, 137(16): 2643–2652.
( 0) 0)
|
| [63] |
GORDON F E, NUTT C L, CHEUNSUCHON P, et al. Increased expression of angiogenic genes in the brains of mouse Meg3-null embryos[J].
Endocrinology, 2010, 151(6): 2443–2452.
( 0) 0)
|
| [64] |
SEITZ H, ROYO H, BORTOLIN M L, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain[J].
Genome Research, 2004, 14(9): 1741–1748.
( 0) 0)
|
| [65] |
余长威.长非编码RNA Gtl2在小鼠胚胎发育早期的表达调控及功能研究[D].硕士学位论文.哈尔滨:哈尔滨工业大学,2014.
( 0) 0)
|
| [66] |
KOPPES E, HIMES K P, CHAILLET J R. Partial loss of genomic imprinting reveals important roles for Kcnq1 and Peg10 imprinted domains in placental development[J].
PLoS One, 2015, 10(8): e0135202.
( 0) 0)
|
| [67] |
宋娜.印记基因PEG10、L3MBTL1在辅助生殖技术出生子代胎盘表达的研究[D].硕士学位论文.郑州:郑州大学,2014.
( 0) 0)
|
| [68] |
CHEN S L, SHI X Y, ZHENG H Y, et al. Aberrant DNA methylation of imprinted H19 gene in human preimplantation embryos[J].
Fertility and Sterility, 2010, 94(6): 2356–2358.
( 0) 0)
|
| [69] |
楼航英.ART对人早孕期胎儿印记基因表达和修饰的影响及其对出生儿血脂代谢改变及相关机制研究[D].博士学位论文.杭州:浙江大学,2013.
( 0) 0)
|
| [70] |
LUX A, BEIL C, MAJETY M, et al. Human retroviral gag-and gag-pol-like proteins interact with the transforming growth factor-β receptor activin receptor-like kinase 1[J].
Journal of Biological Chemistry, 2005, 280(9): 8482–8493.
( 0) 0)
|
| [71] |
PERNA F, GURVICH N, HOYA-ARIAS R, et al. Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells:possible role in 20q-polycythemia vera[J].
Blood, 2010, 116(15): 2812–2821.
( 0) 0)
|
| [72] |
CHOPRA M, AMOR D J, SUTTON L, et al. Russell-Silver syndrome due to paternal H19/IGF2 hypomethylation in a patient conceived using intracytoplasmic sperm injection[J].
Reproductive Biomedicine Online, 2010, 20(6): 843–847.
( 0) 0)
|
| [73] |
RIESEWIJK A M, HU L D, SCHULZ U, et al. Monoallelic expression of human PEG1/MEST is paralleled by parent-specific methylation in fetuses[J].
Genomics, 1997, 42(2): 236–244.
( 0) 0)
|
| [74] |
NAKABAYASHI K, BENTLEY L, HITCHINS M P, et al. Identification and characterization of an imprinted antisense RNA (MESTIT1) in the human MEST locus on chromosome 7q32[J].
Human Molecular Genetics, 2002, 11(15): 1743–1756.
( 0) 0)
|
| [75] |
BELHARAZEM D,KUNANZ J,HENNE A K,et al.Loss of imprinting of insulin-like growth factor-2(IGF2) in colon carcinomas leads to the cell cycle genes activation-hints to intense proliferation[C]//Virchows Archive.New York:Springer,2012,461:S29.
( 0) 0)
|
| [76] |
王颖.异常出生体重胎儿胎盘印记基因PEG1与PEG3的表达与启动子区甲基化及意义[D].硕士学位论文.沈阳:中国医科大学,2010.
( 0) 0)
|
| [77] |
MCMINN J, WEI M, SCHUPF N, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction[J].
Placenta, 2006, 27(6/7): 540–549.
( 0) 0)
|
| [78] |
SHI X, HE Z, GAO Y, et al. Placental expression of PHLDA2 in selective intrauterine growth restriction in monozygotic twins[J].
Placenta, 2014, 35(6): 428–430.
( 0) 0)
|
| [79] |
NAKAMURA T, LIU Y J, NAKASHIMA H, et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos[J].
Nature, 2012, 486(7403): 415–419.
( 0) 0)
|
| [80] |
LIU L Z, MAO S Q, RAY C, et al. Differential regulation of genomic imprinting by TET proteins in embryonic stem cells[J].
Stem Cell Research, 2015, 15(2): 435–443.
( 0) 0)
|




