2. 甘肃农业大学动物科学技术学院, 兰州 730070
2. College of Animal Science and Technology, Gansu Agricultural University, Lanzhou 730070, China
过氧化物酶体增殖物激活受体γ(preoxisome proliferator-activated receptor gamma,PPARγ)属于核受体家族中的一员,只有在被配体激活时才具有转录活性从而在转录水平上调控多种信号通路基因mRNA表达。不仅通过抑制核转录因子-κB(nuclear factor-κB,NF-κB)、c-Jun氨基末端激酶(c-Jun N-terminal kinase,JNK)等炎症通路基因的转录而发挥抗炎作用[1-2],参与单核细胞、巨噬细胞等细胞的增殖、分化和凋亡[3-5],而且PPARγ信号通路还是调控脂肪代谢的必要转录因子,处于脂质沉积信号传递的核心枢纽位置[6-8],通过调控脂肪代谢相关基因的转录促进脂质的沉积[9-11]。在脂肪细胞形成早期起着开关的作用,通过调控脂肪细胞形成、脂肪沉积相关基因的转录参与脂肪代谢全过程。一方面,多能间充质干细胞只有在PPARγ存在的情况下,才有可能分化为脂肪细胞;另一方面,在脂肪细胞增殖分化和成熟的全过程中,参与脂肪代谢的关键基因都受PPARγ信号通路的调控[12]。此外,PPARγ还和脂肪因子的分泌有关[13],脂肪因子通过影响细胞外基质成分,改变脂肪细胞生存微环境,从而通过细胞与微环境之间的相互作用将信号传递给脂肪细胞,细胞胞内PPARγ再通过信号传导调节机体脂肪代谢。本文对PPARγ结构特征、PPARγ调控靶基因特征、PPARγ信号通路对脂质代谢的调控以及PPARγ基因变异对脂质代谢的影响进行了系统阐述,并针对目前脂肪代谢调控亟待解决的一些问题对PPARγ的今后研究重点进行了展望,以期为更深入地探究通过PPARγ信号通路及其靶基因调控脂质代谢提供参考。
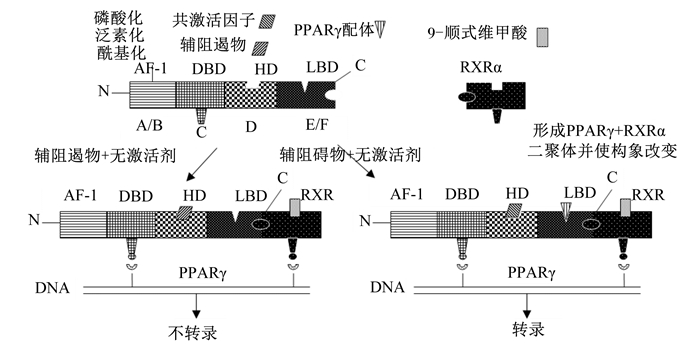
1 PPARγ的结构及调控靶基因的基本特征 1.1 PPARγ的结构PPARγ为维甲酸、类固醇、甲状腺受体超家族中的成员,属于只有在配体存在的情况下才能发挥活性的受配体激活的一类核内转录因子,1990年由英国科学家Issemann和Green首次在大鼠的肝脏组织中克隆鉴定了过氧化物酶体增殖物激活受体α(PPARα)[14],随后从非洲爪蟾、人类和小鼠中分别发现了不同亚基类型的基因过氧化物酶体增殖物激活受体β(PPARβ)、PPARγ,以人类的PPARγ为例,PPARγ含有479个氨基酸残基,和PPARα、PPARβ一样,都包含A、B、C、D、E和F等6个结构域,划分为4个功能区[15-16],见图 1。PPARγ 4个功能区依次为1)N端转录活化区,即A/B区,该区域含有可被磷酸化激活的丝氨酸273(serine 273,ser 273),ser 273氨基酸残基去磷酸化后,促进PPARγ和配体的结合,反之,当ser 273磷酸化后,抑制PPARγ与配体的结合[17];2)DNA结合区(DNA-binding domain,DBD),即C区,该区域含有70多个氨基酸残基,其高度保守的核苷酸序列可以形成2个锌指结构,锌指结构与维甲酸X受体α(retinoid X receptor α,RXRα)结合形成异源二聚体,其构象发生改变与PPARγ配体结合后再与靶基因启动子区的过氧化物酶体增殖反应元件(peroxisome proliferator responsive elements,PPREs)结合,调节靶基因的转录;3)铰链区(hinge domain,HD),即D区,该区域是PPARγ与转录辅助因子互作的区域,通过自身的变形调节PPARγ转录活性;4)配体结合区(ligand-binding domain,LBD),即E/F区,该区域位于PPARγ的羧基端,在PPARγ发挥转录活性中起着关键作用,主要功能是参与PPARγ与配体特异性的结合。另外,与PPARα、PPARβ相比,PPARγ的配体结合区域主要由亲水性氨基酸组成,因而容易与亲水性的配体结合,如不饱和脂肪酸等[18],而PPARα、PPARβ的配体区更容易与亲脂性的饱和脂肪酸结合。

|
PPAR:过氧化物酶体增殖物激活受体preoxisome proliferator-activated receptor。 图 1 PPAR的功能结构域(A)和异构体结构(B)示意图 Fig. 1 Schematic diagram of functional domain (A) and isomer structure (B) of PPAR[16] |
PPAR家族中PPARγ和PPARα、PPARβ在结构上存在差异,因而三者的功能也存在差异,就对脂肪代谢的调节而言,PPARα和PPARβ主要功能是加速脂肪的氧化供能[19],而PPARγ主要功能是诱导脂肪细胞的分化、克隆扩增,促进脂肪的沉积,在脂肪组织中具有较高的表达水平。在发挥转录调节作用时,PPARγ首先与RXRα结合形成异源二聚体,在配体存在的情况下,异源二聚体构象发生改变,然后与靶基因启动子区序列为AGGACAAAGGTCA的PPREs结合,从而通过激活或抑制PPARγ信号通路上一系列脂肪代谢特异性基因mRNA的表达,从而在转录水平上调控脂肪细胞的增殖分化和脂质沉积[20-22],PPARγ的激活转录过程见图 2。
|
AF-1:激活结构域activation function-1;DBD:DNA结合域DNA binding domain;HD:铰链区hinge domain;LBD:配体结合域ligand-binding domain;RXRa:维甲酸X受体α retinoid X receptor α;PPAR:过氧化物酶体增殖物激活受体preoxisome proliferator-activated receptor。 图 2 PPARγ的激活转录过程 Fig. 2 Activation of transcription process of PPARγ[18] |
由此可见,配体在PPARγ发挥转录活性过程中起着至关重要的作用,在缺乏配体的情况下,PPARγ就失去其转录调控活性。PPARγ的配体(激动剂)比较广泛,一类是天然配体,这类配体来源于食物,或者是机体的代谢活动的产物,如来源于食物的一些不饱和脂肪酸(亚油酸、亚麻酸、花生四烯酸等)以及来源于动物机体的代谢产物(前列腺素、氧化低密度脂蛋白等);另一类是人工合成的配体,如罗格列酮、吡格列酮、布洛芬和芳基酪氨酸衍生物等[23-26]。此外,PPARγ的活性还受DNA甲基化、磷酸化、酰基化、泛素化、亚硝基化、硝基化、组蛋白修饰、非编码RNA和染色质重塑等表观修饰的影响[27],表观修饰调控对PPARγ活性的调控可能是今后一段时间内研究的重点。
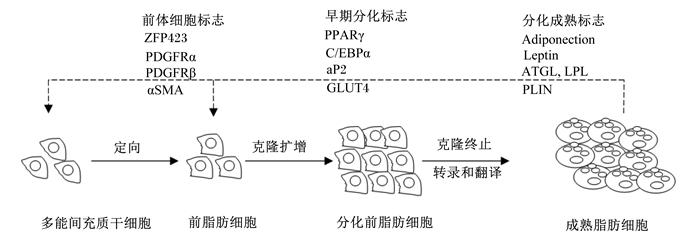
2 PPARγ对脂质合成与代谢的调控 2.1 PPARγ对脂肪细胞形成的调控当动物体摄入的能量大于维持能量时,多余的能量便以脂肪形式储存在体内,当摄入的能量小于维持能量时,体内储存的脂肪被动员供给机体能量,以便维持机体正常的生理活动。所以,动物的脂肪组织是体内最大的能量储存器官,是机体能量平衡的调节器。脂肪的沉积由脂肪细胞数量的增多和体积的增大2方面引起,而脂肪细胞则是由多能间充质干细胞(mesenchymal stem cells,MSC)在成脂因子的刺激下发育而成,要使MSC分化为成熟脂肪细胞必须经过4个阶段[28](图 3)。
|
ZFP423:锌指蛋白423 zinc finger protein 423;PDGFRα:血小板源性生长因子α platelet-derived growth factor receptor α;PDGFRβ:血小板源性生长因子β platelet-derived growth factor receptor β;αSMA:α平滑肌肌动蛋白α smooth muscle actin;PPARγ:过氧化物酶体增殖物激活受体preoxisome proliferator-activated receptor;C/EBPα:CCAAT/增强子结合蛋白α CCAAT/enhancer binding protein α;aP2:脂肪型脂肪酸结合蛋白adipocyte fatty acid binding protein;GLUT4:葡萄糖转运蛋白4 glucose transport protein 4;Adiponection:脂联素;Leptin:瘦素;ATGL:脂肪甘油三酯脂肪酶adipose triglyceride lipase;LPL:脂蛋白脂肪酶lipoprotein lipase;PLIN:围脂滴蛋白perilipin。 图 3 脂肪细胞的形成过程 Fig. 3 Forming process of adipocyte[28] |
第1个阶段:MSC向脂肪细胞定向阶段。MSC在胰岛素、胎牛血清等成脂因子诱导下可定向形成静息前脂肪细胞,此阶段的标志为锌指蛋白423(zinc finger protein 423,ZFP423)[29]、血小板源性生长因子α(platelet-derived growth factor receptor α,PDGFRα)[30]、血小板源性生长因子β(platelet-derived growth factor receptor β,PDGFRβ)[31]、α平滑肌肌动蛋白(α smooth muscle actin,αSMA)[32-33]等。
第2个阶段:克隆扩增阶段。定向的前脂肪细胞在胰岛素样生长因子等的诱导下克隆扩增,形成分化前脂肪细胞,此阶段标志为PPARγ[34-37]、CCAAT/增强子结合蛋白α(CCAAT/enhancer binding protein,C/EBPα)[38]、脂肪型脂肪酸结合蛋白(adipocyte fatty acid binding protein,aP2)[39]、葡萄糖转运蛋白4(glucose transport protein 4,GLUT4)[34]等。
第3个阶段:克隆终止阶段。在PPARγ和C/EBPα的调控下,前脂肪细胞克隆扩增,脂肪细胞数量增多,为脂肪沉积创造条件,当克隆扩增达到一定程度,在C/EBPα的诱导下,前脂肪细胞克隆终止[40],为细胞进入终末分化创造条件。
第4个阶段:转录和翻译阶段。此阶段是脂肪细胞分化的终末阶段,经过转录和翻译脂肪细胞分化为最终的成熟脂肪细胞,此阶段的标志为脂联素(adiponection)、瘦素(leptin)、脂肪甘油三酯脂肪酶(adipose triacylglyceride lipase,ATGL)、脂蛋白脂肪酶(lipoprteinlipase,LPL)、围脂滴蛋白(perilipin,PLIN)等[41-45],这些标志基因都为PPARγ信号通路的靶基因[12],PPARγ通过调节这些标志性靶基因的转录参与能量代谢和脂质沉积,诱导脂肪细胞的分化和增殖,促进脂肪酸的转运和沉积,参与甘油三酯的形成和脂解的生化循环,进而影响脂肪代谢和脂肪沉积[46]。
目前的研究表明,MSC只有在PPARγ存在的情况下,才有可能向脂肪细胞分化,然而PPARγ如何开启MSC向脂肪细胞的分化以及相关的信号传导机制还不明确,随着生物科学的发展和研究的深入,PPARγ与脂肪细胞之间的调控关系会进一步揭示。
2.2 PPARγ对脂质代谢的调控脂肪沉积是受PPARs、C/EBPs、固醇调节元件结合蛋白(sterol-regulatory element binding proteins,SREBPs)和环磷酸腺苷/蛋白激酶A(cyclic adenosine 3′, 5′ monophosphate/ protein kinase A,cAMP/PKA)等多种信号通路调控的复杂生物过程[47-48]。其中PPARγ信号通路是调控脂肪代谢的主要信号通路,处于信号通路调控网络的核心枢纽位置,参与脂肪代谢、葡萄糖代谢、胰岛素敏感等重要生物过程[49-50],在脂肪细胞的基因表达、脂肪细胞与细胞微环境之间的信号传导以及脂肪细胞之间信息传递过程中扮演着主要调控器的作用。PPARγ在脂肪、肌肉、内脏器官等组织中均有表达,脂肪中表达量最高[51],与C/EBPα或C/EBPβ相互促进表达调节脂肪细胞分化与脂肪沉积[21]。有研究证实,PPARγ不但能诱导MSC分化为前脂肪细胞,还能使成对数生长期纤维细胞转化为脂肪细胞[52]。
PPARγ可调控一系列脂肪代谢特异性基因的表达[53-55],在有配体存在的情况下,通过与下游靶基因脂肪酸合成酶(fatty acid synthase,FAS)、硬脂酰辅酶A去饱和酶(stearoy-CoA desaturase,SCD)、苹果酸酶(malic enzyme,ME)、脂肪酸结合蛋白(fatty acid-binding protein,FABP)、脂蛋白脂肪酶(lipoprteinlipase,LPL)、乙酰辅酶A合成酶(acetyl-CoA synthetase,ACS)、脂肪酸转运蛋白(fat acid transport proteins,FATP)、脂滴包被蛋白(perilipin,PLIN)、磷酸烯醇式丙酮酸羧激酶(phosphoenolpyruvate carboxykinase,PEPCK)等启动子区的PPREs结合调控其表达[12, 52, 56],从而参与脂肪合成、转运和沉积等脂肪代谢的全过程[12, 52, 56]。PPARγ通过调控FAS、SCD等脂肪合成相关基因的表达,参与脂肪的合成;通过调控FATP、A-FABP等脂肪转运相关基因的表达,促进脂肪酸的转运和吸收;通过介导肉碱棕榈酰易位酶(carnitine acetyltranslocase,CPT)、中链脂酰辅酶A脱氢酶(medium chain acyl-CoA dehydrogenase,MCAD)、长链脂酰辅酶A脱氢酶(long chain acyl-CoA dehydrogenase,LCAD)等的表达调节脂肪酸的β氧化[57-60],参与甘油三酯的形成和脂解的生化循环[61],进而调节脂肪代谢[46];通过与瘦素调控元件结合,调控瘦素基因的表达维持脂肪相对稳态[62]的研究显示,用PPARγ配体吡格列酮分别作用于人胎盘滋养层细胞和山羊乳腺后,脂肪代谢相关基因FABP4、PLIN2[63]、LPL、FAS、FABP3和SCD等[64]上调,促进脂肪沉积。PPARγ还可促进小鼠肝脏FATP、FAS、aP2、LPL等的表达,促进脂肪细胞的增大和脂质沉积,抑制PPARγ表达时,可减少小鼠脂肪沉积量和白色脂肪细胞的大小,降低血清低密度脂蛋白(low density lipoprotein,LDL)、葡萄糖(glucose,GLU)的水平[65]。在对莱芜猪、鲁莱黑猪和大约克夏猪3个不同品种猪的背最长PPARγ表达量和肌内脂肪含量的研究中发现,不同品种猪的背最长肌PPARγ表达量有显著品种差异,但都和肌内脂肪含量呈显著正相关[66]。携带有徐淮山羊PPARγ基因的湖羊背最长肌肌内脂肪含量与对照组相比,显著提高了7.8个百分点[67]。Liu等[68]也报道,PPARγ的mRNA表达量和肌内脂肪含量呈显著正相关,PPARγ可以作为肉品质性状的主要候选基因[69]。此外,PPARγ还是血糖、血脂稳态的重要因子[70],PPARγ缺乏导致胰岛素敏感性增加[71]。这些研究结果表明,PPARγ基因是肌内脂肪含量的主要影响因子,在脂肪沉积过程中发挥重要作用,可能是今后肉品质调控研究的一个重要基因。
PPARγ在脂肪细胞的定向形成过程中起着开关的作用,至今尚未发现细胞可以在PPARγ缺失的情况下向脂肪细胞分化。将小鼠胚胎干细胞中的PPARγ基因沉默处理后,小鼠胚胎干细胞能分化为肌肉细胞、神经细胞、造血干细胞等,但唯独不能形成脂肪细胞[72]。用PPARγ抑制剂沉默信息调节因子(silent information regulator 1,SIRT1)抑制小鼠白色脂肪中的PPARγ时,脂肪结合蛋白表达降低,脂肪动员增加[73],反之,敲除小鼠脂肪组织中的SIRT1时,脂肪细胞增大,脂肪沉积增加。在人上的研究显示,PPARγ基因Pro115Gln突变会导致PPARγ的活性增强,表现为显著肥胖,而Phe388Leu、Pro467Leu、Arg425Cys的突变引起脂质代谢障碍、脂肪转移[74]。体外试验表明,在没有C/EBPα存在的情况下,PPARγ仍可促进脂肪细胞的分化,但反过来只有C/EBPα存在的情况下MSC不能分化为脂肪细胞[75]。这说明只有在转录因子PPARγ存在时,MSC才有可能向脂肪细胞分化。由此可见,PPARγ通过调节其通路上脂肪代谢关键的靶基因的表达,从mRNA水平上调节脂肪细胞的增殖、分化和脂肪积累,是脂质代谢和能量代谢的关键调节因子,PPARγ在蛋白水平上的调节机制及靶向介导的生物学效应可能是今后一段时间内研究的焦点。
2.3 PPARγ对脂肪因子的调控脂肪不仅是机体内的能量储存器,还是重要的内分泌系统之一,通过分泌瘦素、脂联素、前列腺素、抗肿瘤坏死因子-α、白细胞介素-6、血管内皮生长因子等[76-77]调节机体的能量代谢。已有研究结果显示,PPARγ与脂肪因子的分泌有关,PPARγ不仅通过抑制Janus激酶/信号转导与转录激活子信号通路基因的表达,从而抑制瘦素的合成[78-79],而且通过调节脂联素启动子反应元件促进脂联素合成[80]。在小鼠上的研究结果表明,PPARγ配体可以促进高脂诱导的小鼠内脂素[81]、抵抗素[82]分泌增多。也有研究表明,PPARγ转录活性与抵抗素释放呈相反关系[83]。因此,转录因子PPARγ与脂肪因子之间的关系还需要进一步研究明确。
3 营养水平通过PPARγ调节脂肪沉积营养水平从转录水平上调控PPARγ的表达,目前,营养水平对PPARγ的调控主要集中在饲粮能量和蛋白质。高能量饲粮促进黄牛和西门塔尔杂交肉牛皮下脂肪组织PPARγ的表达[84],促进皮下脂肪、内脏脂肪沉积。高脂饲粮可使肉犊牛背最长肌PPARγ的mRNA表达量显著增大,肌内脂肪沉积量增加,改善牛肉品质[85]。在羊上的研究结果也显示,摄入高能量饲粮时,杂交肉羊背最长肌、皮下脂肪、尾部脂肪等组织的PPARγ表达量显著升高,肌内脂肪含量和各部位脂肪沉积量增加[58, 86]。高蛋白质饲粮对PPARγ的表达有抑制作用,用高蛋白质饲粮饲喂的乌金猪脂肪组织中PPARγ的表达显著降低[87]。高蛋白质水平提高动物机体的热增耗,从而导致机体能量利用效率降低,故蛋白质水平可能也是通过能量途径抑制PPARγ的表达[86]。营养水平是除了遗传、环境外,影响动物机体脂肪沉积和肉品质的最重要的因素,通过营养调控肉品质以及能量、蛋白质、维生素等各营养物质之间的相互作用在现代动物生产中具有举足轻重的作用。
4 PPARγ与生物钟相互反馈调控脂质沉积机体能量代谢、脂质代谢与生物钟密切相关。研究表明,小鼠肝脏中PPARγ的抑制剂SIRT1具有维持生物钟相关基因稳定表达的作用,敲除SIRT1基因后,生物钟基因脑肌类芳香烃受体转位蛋白1(muscle aryl hydrocarbon receptor nuclear translocator-like protein 1,BMAL1)、昼夜节律运动的输出故障(circadian locomotor output cycles kaput,CLOCK)的mRNA和蛋白表达量均下降[88],而且BMAL1、CLOCK可以和SIRT1形成复合体调控生物钟相关基因和PPARγ的表达。PPARγ不仅是脂肪代谢的关键转录因子,还是调节生物钟的候选基因之一[89],高脂肪饮食小鼠肝脏中PPARγ及其配体亚麻酸、花生四烯酸表达量升高[90],生物钟基因核受体亚家族1表达量降低[91],引起肥胖,这种高脂饮食通过激活PPARγ周期变化干扰正常生理节律的现象与饮食引起的肥胖无关[92]。一方面,生物钟调控能量代谢,生物钟基因BMAL1-CLOCK可以和PPARγ的抑制剂SIRT1形成复合体调控PPARγ的表达,PPARγ共激活因子α的活性也受生物钟调控。另一方面,能量代谢反过来又调节生物钟,PPARγ共激活因子α、PPARγ抑制剂SIRT1也可以调节生物钟。PPARγ与生物钟相互反馈调控生物钟和脂肪沉积,生物钟的紊乱除了会造成脂肪代谢异常,引起肥胖外,还会引起心血管疾病、心脏病等一系列并发症的发生[93]。这种PPARγ与生物钟相互反馈调节的关系在脂质代谢中发挥重要作用,可能会成为治疗肥胖和代谢并发症的新目标。
5 小结与展望综上所述,PPARγ作为一种多效调控工具,参与多种细胞的增殖分化,不仅在转录水平上调控脂肪细胞增殖分化和脂质沉积,而且PPARγ与生物钟相互反馈调控脂质代谢。近年来,随着人民生活水平的提高和人们饮食结构的改变,对肉的嫩度、多汁性、大理石纹等提出了更高的要求,生产适宜肌内脂肪含量的肉品是现代畜牧业生产者和研究者普遍关注的焦点问题之一。肌内脂肪不仅能保证肉品的嫩度和多汁性,而且能改善肌肉的系水力和嫩度,如何通过有效地调控PPARγ控制肉品质,生产符合消费者期望的畜禽产品具有重大意义。关于PPARγ对脂肪代谢调节的分子机制,尽管已进行了大量的系统研究,也取得了一些重大进展,但还存在许多问题,诸如PPARγ如何开启MSC向脂肪细胞的分化,PPARγ相关信号通路的传导机制以及PPARγ与脂肪因子瘦素、脂联素、抵抗素等的相互激活转录机制,特别是如何通过有效地干预PPARγ的转录调控脂肪代谢靶向控制PPARγ介导的生物学效应,今后需对这些问题进一步深入探索,以期为通过PPARγ调控畜禽脂质代谢和提高肉品质提供参考。
| [1] |
RANI N, BHARTI S, BHATIA J, et al. Chrysin, a PPAR-γ agonist improves myocardial injury in diabetic rats through inhibiting AGE-RAGE mediated oxidative stress and inflammation[J]. Chemico-Biological Interactions, 2016, 250: 59-67. DOI:10.1016/j.cbi.2016.03.015 |
| [2] |
MOTOKI T, KUROBEE H, HIRATA Y, et al. PPAR-γ agonist attenuates inflammation in aortic aneurysm patients[J]. General Thoracic and Cardiovascular Surgery, 2015, 63(10): 565-571. DOI:10.1007/s11748-015-0576-1 |
| [3] |
VICTOR N A, WANDERI E W, GAMBOA J, et al. Altered PPARγ expression and activation after transient focal ischemia in rats[J]. European Journal of Neuroscience, 2006, 24(6): 1653-1663. DOI:10.1111/j.1460-9568.2006.05037.x |
| [4] |
AL ROUQ F, EL ETER E. PPAR-γ activator induces neuroprotection in hypercholesterolemic rats subjected to global cerebral ischemia/reperfusion injury:in vivo and in vitro inhibition of oxidative stress[J]. Experimental Gerontology, 2014, 51: 1-7. DOI:10.1016/j.exger.2013.12.008 |
| [5] |
KADAM L, KILBURN B, BACZYK J, et al. Rosiglitazone blocks first trimester in-vitro placental injury caused by NF-κB-mediated inflammation[J]. Science Report, 2019, 9: 2018. DOI:10.1038/s41598-018-38336-2 |
| [6] |
TOMAS J, MULET C, SAFFARIAN A, et al. High-fat diet modifies the PPAR-γ pathway leading to disruption of microbial and physiological ecosystem in murine small intestine[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(40): E5934-E5943. DOI:10.1073/pnas.1612559113 |
| [7] |
MULLER E, DRPRI S, AIYER A, et al. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor γ isoforms[J]. Journal of Biological Chemistry, 2002, 277(44): 41925-41930. DOI:10.1074/jbc.M206950200 |
| [8] |
RESNYK C W, CARRÉ W, WANG X F, et al. Transcriptional analysis of abdominal fat in chickens divergently selected on bodyweight at two ages reveals novel mechanisms controlling adiposity:validating visceral adipose tissue as a dynamic endocrine and metabolic organ[J]. BMC Genomics, 2017, 18: 626. DOI:10.1186/s12864-017-4035-5 |
| [9] |
XU L Y, MA X R, VERMA N, et al. Ablation of PPARγ in subcutaneous fat exacerbates age-associated obesity and metabolic decline[J]. Aging Cell, 2018, 17(2): e12721. |
| [10] |
白祥, 禹宝庆, 黄建明, 等. PPARγ基因干扰体外对兔骨髓间充质干细胞成脂基因的影响[J]. 中国老年学杂志, 2018, 38(18): 4478-4481. DOI:10.3969/j.issn.1005-9202.2018.18.046 |
| [11] |
MOISÁ J M, SHIKE D W, FAULKNER D B, et al. Central role of the PPARγ gene network in coordinating beef cattle intramuscular adipogenesis in response to weaning age and nutrition[J]. Gene Regulation and Systems Biology, 2014, 8: 17-32. |
| [12] |
YANG H, SUH D H, KIM E D, et al. Metabolomic and lipidomic analysis of the effect of pioglitazone on hepatic steatosis in a rat model of obese type 2 diabetes[J]. British Journal of Pharmacology, 2018, 175(17): 3610-3625. DOI:10.1111/bph.14434 |
| [13] |
ELISSA L A, ELSHERBINY N M, MAGMOMAH A O. Propolis restored adiponectin level in type 2 diabetes through PPARγ activation[J]. Egyptian Journal of Basic and Applied Sciences, 2015, 2(4): 318-326. DOI:10.1016/j.ejbas.2015.06.003 |
| [14] |
ISSEMANN I, GREEN S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators[J]. Nature, 1990, 347(6294): 645-650. DOI:10.1038/347645a0 |
| [15] |
LEE W S, KIM J. Peroxisome proliferator-activated receptors and the heart:lessons from the past and future directions[J]. PPAR Research, 2015, 2015: 271983. |
| [16] |
USUDA D, KANDA T. Peroxisome proliferator-activated receptors for hypertension[J]. World Journal of Cardiology, 2014, 6(8): 744-754. DOI:10.4330/wjc.v6.i8.744 |
| [17] |
杨谷良, 潘敏雄, 向福, 等. PPARγ调控脂肪细胞增殖和分化机理研究进展[J]. 食品科学, 2017, 38(3): 254-260. |
| [18] |
HSU M H, PALMER C N A, SONG W, et al. A carboxyl terminal extension of the zinc finger domain contributes to the specificity and polarity of peroxisome proliferator-activated receptor DNA binding[J]. Journal of Biological Chemistry, 1998, 273(43): 27988-27997. DOI:10.1074/jbc.273.43.27988 |
| [19] |
BARROSO E, RODRÍGUEZ-CALVO R, SERRANO-MARCO L, et al. The PPARβ/δ activator GW501516 prevents the down-regulation of AMPK caused by a high-fat diet in liver and amplifies the PGC-1α-Lipin 1-PPARα pathway leading to increased fatty acid oxidation[J]. Endocrinology, 2011, 152(5): 1848-1859. DOI:10.1210/en.2010-1468 |
| [20] |
TAKAHASHI S, TANAKA T, KODAMA T, et al. Peroxisome proliferator-activated receptor δ (PPARδ), a novel target site for drug discovery in metabolic syndrome[J]. Pharmacological Research, 2006, 53(6): 501-507. DOI:10.1016/j.phrs.2006.03.019 |
| [21] |
JIA Y Y, WU C Y, KIM J, et al. Astaxanthin reduces hepatic lipid accumulations in high-fat-fed C57BL/6J mice via activation of peroxisome proliferator-activated receptor (PPAR) alpha and inhibition of PPAR gamma and Akt[J]. Journal of Nutritional Biochemistry, 2016, 28: 9-18. DOI:10.1016/j.jnutbio.2015.09.015 |
| [22] |
NUNN A V, BELL J, BARTER P. The integration of lipid-sensing and anti-inflammatory effects:how the PPARs play a role in metabolic balance[J]. Nuclear Receptor, 2007, 5(1): 1. DOI:10.1186/1478-1336-5-1 |
| [23] |
GARCIA-VALLVÉ S, GUASCH L, TOMAS-HERNÁNDEZ S, et al. Peroxisome proliferator-activated receptor γ(PPARγ) and ligand choreography:newcomers take the stage[J]. Journal of Medicinal Chemistry, 2015, 58(14): 5381-5394. DOI:10.1021/jm501155f |
| [24] |
MALTAROLLO V G, KRONENBERGER T, WINDSHUGEL B, et al. Advances and challenges in drug design of PPARδ ligands[J]. Current Drug Targets, 2018, 19(2): 144-154. DOI:10.2174/1389450118666170414113159 |
| [25] |
YI W, SHI J J, ZHAO G G, et al. Identification of a novel selective PPARγ ligand with a unique binding mode and improved therapeutic profile in vitro[J]. Scientific Reports, 2017, 7: 41487. DOI:10.1038/srep41487 |
| [26] |
IM C N. Combination treatment with PPARγ ligand and its specific inhibitor GW9662 downregulates BIS and 14-3-3 gamma, inhibiting stem-Like properties in glioblastoma cells[J]. BioMed Research International, 2017, 2017: 5832824. |
| [27] |
SHOAITO H, PETIT J, CHISSEY A, et al. The role of peroxisome proliferator-activated receptor gamma (PPARγ) in mono (2-ethylhexyl) phthalate (MEHP)-mediated cytotrophoblast differentiation[J]. Environmental Health Perspectives, 2019, 127(2): 027003. DOI:10.1289/EHP3730 |
| [28] |
GHABEN A L, SCHERER P E. Adipogenesis and metabolic health[J]. Nature Reviews Molecular Cell Biology, 2019, 20(4): 242-258. DOI:10.1038/s41580-018-0093-z |
| [29] |
SHAO M, HEPLER C, VISHVANATH L, et al. Fetal development of subcutaneous white adipose tissue is dependent on Zfp423[J]. Molecular Metabolism, 2017, 6(1): 111-124. |
| [30] |
SHIRASAWA H, HORIUCHI K, OKI S, et al. Targeting of PDGFRα suppresses fat infiltration after rotator cuff tear[J]. Journal of Shoulder and Elbow Surgery, 2017, 26(4): e112. |
| [31] |
OLSON L E, SORIANO P. PDGFRβ signaling regulates mural cell plasticity and inhibits fat development[J]. Developmental Cell, 2011, 20(6): 815-826. DOI:10.1016/j.devcel.2011.04.019 |
| [32] |
FLORIAN R, GENEVIEVE D, CHRISTINA C, et al. Human intestinal mesenchymal cells exhibit a pro-fibrogenic phenotype in response to adipose tissue derived-mediators-linking fat to intestinal fibrosis[J]. Inflammatory Bowel Diseases, 2014, 20: S105. |
| [33] |
CÔTÉ J A, LESSARD J, PELLETIER M, et al. Role of the TGF-β pathway in dedifferentiation of human mature adipocytes[J]. FEBS Open Bio, 2017, 7(8): 1092-1101. DOI:10.1002/2211-5463.12250 |
| [34] |
FERNYHOUGH M E, OKINE E, HAUSMAN G, et al. PPARγ and GLUT-4 expression as developmental regulators/markers for preadipocyte differentiation into an adipocyte[J]. Domestic Animal Endocrinology, 2007, 33(4): 367-378. DOI:10.1016/j.domaniend.2007.05.001 |
| [35] |
BOUABOULA M, HILAIRET S, MARCHAND J, et al. Anandamide induced PPARγ transcriptional activation and 3T3-L1 preadipocyte differentiation[J]. European Journal of Pharmacology, 2005, 517(3): 174-181. DOI:10.1016/j.ejphar.2005.05.032 |
| [36] |
WON P K, WAKI H, CHOI S P, et al. The small molecule phenamil is a modulator of adipocyte differentiation and PPARγ expression[J]. Journal of Lipid Research, 2010, 51(9): 2775-2784. DOI:10.1194/jlr.M008490 |
| [37] |
KANG S U, KIM H J, KIM D H, et al. Nonthermal plasma treated solution inhibits adipocyte differentiation and lipogenesis in 3T3-L1 preadipocytes via ER stress signal suppression[J]. Scientific Reports, 2018, 8: 2277. DOI:10.1038/s41598-018-20768-5 |
| [38] |
LUO X, LI H X, YANG G S. Sequential expression of Wnt/β-catenin signal pathway related genes and adipocyte transcription factors during porcine adipose tissue development[J]. Chinese Journal of Biotechnology, 2008, 24(5): 746-753. DOI:10.1016/S1872-2075(08)60039-4 |
| [39] |
HE J, TIAN Y, LI J J, et al. Expression pattern of adipocyte fatty acid-binding protein gene in different tissues and its regulation of genes related to adipocyte differentiation in duck[J]. Poultry Science, 2012, 91(9): 2270-2274. DOI:10.3382/ps.2012-02149 |
| [40] |
王启贵, 李辉, 张富春. C/EBPs与脂肪细胞的分化调控[J]. 黑龙江畜牧兽医, 2007(12): 19-20. DOI:10.3969/j.issn.1004-7034.2007.12.006 |
| [41] |
CÓTÉ J A, GUÉNARD F, LESSARD J, et al. Temporal changes in gene expression profile during mature adipocyte dedifferentiation[J]. International Journal of Genomics, 2017, 2017: 5149362. |
| [42] |
ONO H, OKI Y, BONO H, et al. Gene expression profiling in multipotent DFAT cells derived from mature adipocytes[J]. Biochemical and Biophysical Research Communications, 2011, 407(3): 562-567. DOI:10.1016/j.bbrc.2011.03.063 |
| [43] |
WEI S J, DUARTE M S, ZAN L S, et al. Cellular and molecular implications of mature adipocyte dedifferentiation[J]. Journal of Genomics, 2013, 1: 5-12. DOI:10.7150/jgen.3769 |
| [44] |
CAT A N D, BRIONES A M.Isolation of mature adipocytes from white adipose tissue and gene expression studies by real-time quantitative RT-PCR[M]//TOUYZ R, SCHIFFRIN E.Hypertension.New York, NY: Humana Press, 2017, 1527: 283-295.
|
| [45] |
MAURIZI G, PETÄISTÖ T, MAURIZI A, et al. Key-genes regulating the liposecretion process of mature adipocytes[J]. Journal of Cellular Physiology, 2017, 233(5): 3784-3793. DOI:10.1002/jcp.26188 |
| [46] |
KRALISCH S, FASSHAUER M. Adipocyte fatty acid binding protein:a novel adipokine involved in the pathogenesis of metabolic and vascular disease?[J]. Diabetologia, 2013, 56(1): 10-21. DOI:10.1007/s00125-012-2737-4 |
| [47] |
ROSEN E D, MACDOUGALD O A. Adipocyte differentiation from the inside out[J]. Nature Reviews Molecular Cell Biology, 2006, 7(12): 885-896. DOI:10.1038/nrm2066 |
| [48] |
ROSEN E D, SPIEGELMAN B M. Molecular regulation of adipogenesis[J]. Annual Review of Cell and Developmental Biology, 2000, 16: 145-171. DOI:10.1146/annurev.cellbio.16.1.145 |
| [49] |
BROWN J D, PLUTZKY J. Peroxisome proliferator-activated receptors as transcriptional nodal points and therapeutic targets[J]. Circulation, 2007, 115(4): 518-533. DOI:10.1161/CIRCULATIONAHA.104.475673 |
| [50] |
COSTA V, FOTI D, PAONESSA F, et al. The insulin receptor:a new anticancer target for peroxisome proliferator-activated receptor-γ (PPAR-γ) and thiazolidinedione PPARγ agonists[J]. Endocrne Related Cancer, 2008, 15(1): 325-335. DOI:10.1677/ERC-07-0226 |
| [51] |
宋淑珍.能量限制对绵羊脂肪沉积的影响及其机理研究[D].博士学位论文.兰州: 甘肃农业大学, 2017. http://cdmd.cnki.com.cn/Article/CDMD-10733-1018973163.htm
|
| [52] |
TONTONOZ P, HU E, DEVUNE J, et al. PPAR gamma 2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene[J]. Molecular and Cellular Biology, 1995, 15(1): 351-357. DOI:10.1128/MCB.15.1.351 |
| [53] |
TAMORI Y, MASUGI J, NISHINO N, et al. Role of peroxisome proliferator-activated receptor-γ in maintenance of the characteristics of mature 3T3-L1 adipocytes[J]. Diabetes, 2002, 51(7): 2045-2055. DOI:10.2337/diabetes.51.7.2045 |
| [54] |
SIERSBAEK R, NIELSEN R, MANDRUP S. PPARγ in adipocyte differentiation and metabolism-novel insights from genome-wide studies[J]. FEBS Letters, 2010, 584(15): 3242-3249. DOI:10.1016/j.febslet.2010.06.010 |
| [55] |
DARLINGTON G J, ROSS S E, MACDOUGALD O A. The role of C/EBP genes in adipocyte differentiation[J]. Journal of Biological Chemistry, 1998, 273(46): 30057-30060. DOI:10.1074/jbc.273.46.30057 |
| [56] |
GLORIAN M, DUPLUS E, BEALE E G, et al. A single element in the phosphoenolpyruvate carboxykinase gene mediates thiazolidinedione action specifically in adipocytes[J]. Biochimie, 2001, 83(10): 933-943. DOI:10.1016/S0300-9084(01)01343-8 |
| [57] |
ITO K, SUDA T. Metabolic requirements for the maintenance of self-renewing stem cells[J]. Nature Reviews Molecular Cell Biology, 2014, 15(4): 243-256. DOI:10.1038/nrm3772 |
| [58] |
CUI H, KONG Y H, ZHANG H. Oxidative stress, mitochondrial dysfunction, and aging[J]. Journal of Receptor and Signal Transduction Research, 2012, 2012: 646354. |
| [59] |
WANG Y X, LEE C H, TIEP S, et al. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity[J]. Cell, 2003, 113(2): 159-170. DOI:10.1016/S0092-8674(03)00269-1 |
| [60] |
DJOUADI F, AUBEY F, SCHLEMMER D, et al. Peroxisome proliferator activated receptor δ (PPARδ) agonist but not PPARα corrects carnitine palmitoyl transferase 2 deficiency in human muscle cells[J]. The Journal of Clinical Endocrinology & Metabolism, 2005, 90(3): 1791-1797. |
| [61] |
GUAN H P, LI Y, JENSEN M V, et al. A futile metabolic cycle activated in adipocytes by antidiabetic agent[J]. Nature Medicine, 2002, 8(10): 1122-1128. DOI:10.1038/nm780 |
| [62] |
ZHANG Y X, DALLNER O S, NAKADAI T, et al. A noncanonical PPARγ/RXRα-binding sequence regulates leptin expression in response to changes in adipose tissue mass[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(26): E6039-E6047. DOI:10.1073/pnas.1806366115 |
| [63] |
EL DAIRI R, HUUSKONEN P, PASANEN M, et al. Peroxisome proliferator activated receptor gamma (PPAR-γ) ligand pioglitazone regulated gene networks in term human primary trophoblast cells[J]. Reproductive Toxicology, 2018, 81: 99-107. DOI:10.1016/j.reprotox.2018.07.077 |
| [64] |
SHI H B, LUO J, ZHU J J, et al. PPARγ regulates genes involved in triacylglycerol synthesis and secretion in mammary gland epithelial cells of dairy goats[J]. PPAR Research, 2013, 2013: 310948. |
| [65] |
ZHANG Y, FAN S J, HU N, et al. Rhein reduces fat weight in db/db mouse and prevents diet-induced obesity in C57Bl/6 mouse through the inhibition of PPARγ signaling[J]. PPAR Research, 2012, 2012: 374936. |
| [66] |
CUI J X, CHEN W, LIU J, et al. Study on quantitative expression of PPARγ and ADRP in muscle and its association with intramuscular fat deposition of pig[J]. SpringerPlus, 2016, 5: 1501. DOI:10.1186/s40064-016-3187-0 |
| [67] |
朱才业, 李伟, 朱睿, 等. 睾丸注射法制备携带徐淮山羊过氧化氢酶体激活增殖受体γ基因转基因羊的研究[J]. 中国畜牧兽医, 2013, 40(6): 153-157. DOI:10.3969/j.issn.1671-7236.2013.06.034 |
| [68] |
LIU C D, SHEN L Y, DU J Q, et al. The effect of lipid metabolism-related genes on intramuscular fat content and fatty acid composition in multiple muscles[J]. Animal Production Science, 2018, 58(11): 2003-2010. DOI:10.1071/AN16292 |
| [69] |
MA J J, CHAI J, SHANG Y Y, et al. Swine PPAR-γ2 expression upregulated in skeletal muscle of transgenic mice via the swine myozenin-1 gene promoter[J]. Transgenic Research, 2015, 24(3): 409-420. DOI:10.1007/s11248-014-9849-1 |
| [70] |
BALAKRISHNAN B B, KRISHNASAMY K, MAYAKRISHNAN V, et al. Moringa concanensis Nimmo extracts ameliorates hyperglycemia-mediated oxidative stress and upregulates PPARγ and GLUT4 gene expression in liver and pancreas of streptozotocin-nicotinamide induced diabetic rats[J]. Biomedicine & Pharmacotherapy, 2019, 112: 108688. DOI:10.1016/j.biopha.2019.108688 |
| [71] |
CAGLAYAN E, TRAPPIEL M, BEHRINGER A, et al. Pulmonary arterial remodelling by deficiency of peroxisome proliferator-activated receptor-γ in murine vascular smooth muscle cells occurs independently of obesity-related pulmonary hypertension[J]. Respiratory Research, 2019, 20: 42. DOI:10.1186/s12931-019-1003-4 |
| [72] |
ROSEN E D, SARRAF A E, TROY G, et al. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro[J]. Molecular Cell, 1999, 4(4): 611-617. DOI:10.1016/S1097-2765(00)80211-7 |
| [73] |
QIANG L, WANG L H, KON N, et al. Brown remodeling of white adipose tissue by Sirt1-dependent deacetylation of PPARγ[J]. Cell, 2012, 150(3): 620-632. DOI:10.1016/j.cell.2012.06.027 |
| [74] |
王娜, 张志文. PPARγ调节与糖脂质代谢[J]. 生理科学进展, 2005, 36(3): 240. |
| [75] |
TANNKA T, YOSHIDA N, KISHIMOTO T, et al. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene[J]. EMBO Journal, 1997, 16(24): 7432-7443. DOI:10.1093/emboj/16.24.7432 |
| [76] |
FIELDS D A, SCHNEIDER C R, PAVELA G. A narrative review of the associations between six bioactive components in breast milk and infant adiposity[J]. Obesity, 2016, 24(6): 1213-1221. DOI:10.1002/oby.21519 |
| [77] |
SLUTSKY N, VATARESCU M, HAIM Y, et al. Decreased adiponectin links elevated adipose tissue autophagy with adipocyte endocrine dysfunction in obesity[J]. International Journal of Obesity, 2016, 40(6): 912-920. DOI:10.1038/ijo.2016.5 |
| [78] |
AL-JADA D N, AHMAD M N. Dietary fat and insulin resistance:a connection through leptin and PPARγ activation[J]. Functional Foods in Health & Disease, 2016, 6(6): 306-328. |
| [79] |
MYNATT R L, STEPHENS J M. Agouti regulates adipocyte transcription factors[J]. American Journal of Physiology-Cell Physiology, 2001, 280(4): C954-C961. DOI:10.1152/ajpcell.2001.280.4.C954 |
| [80] |
CHINETTI G, ZAWADSKI C, FRUCHART J C, et al. Expression of adiponectin receptors in human macrophages and regulation by agonists of the nuclear receptors PPARα, PPARγ, and LXR[J]. Biochemical and Biophysical Research Communications, 2004, 314(1): 151-158. DOI:10.1016/j.bbrc.2003.12.058 |
| [81] |
OLIVEIRA L S C, SANTOS D A, BARBOSA-DA-SILVA S, et al. The inflammatory profile and liver damage of a sucrose-rich diet in mice[J]. The Journal of Nutritional Biochemistry, 2014, 25(2): 193-200. DOI:10.1016/j.jnutbio.2013.10.006 |
| [82] |
STEPPAN C M, BAILEY S T, BHAT S, et al. The hormone resistin links obesity to diabetes[J]. Nature, 2001, 409(6818): 307-312. DOI:10.1038/35053000 |
| [83] |
WAY J M, GÖRGUN C Z, TONG Q, et al. Adipose tissue resistin expression is severely suppressed in obesity and stimulated by peroxisome proliferator-activated receptor γ agonists[J]. Journal of Biological Chemistry, 2001, 276(28): 25651-25653. DOI:10.1074/jbc.C100189200 |
| [84] |
ZHANG H B, ZHANG X F, WANG Z S, et al. Effects of dietary energy level on lipid metabolism-related gene expression in subcutaneous adipose tissue of yellow breed×Simmental cattle[J]. Animal Science Journal, 2015, 86(4): 392-400. DOI:10.1111/asj.12316 |
| [85] |
SEGERS J R, LOOR J J, MOISÁ S J, et al. Effects of protein and fat concentration in coproduct-based growing calf diets on adipogenic and lipogenic gene expression, blood metabolites, and carcass composition[J]. Journal of Animal Science, 2017, 95(6): 2767-2781. DOI:10.2527/jas.2017.1446 |
| [86] |
王小芳, 曾洁, 田崇奇, 等. 日粮能量和蛋白水平对滩羊脂肪组织PPARγ和FABP4 mRNA表达的影响[J]. 畜牧兽医学报, 2016, 47(11): 2266-2273. DOI:10.11843/j.issn.0366-6964.2016.11.014 |
| [87] |
ZHAO S M, WANG J, SONG X L, et al. Impact of dietary protein on lipid metabolism-related gene expression in porcine adipose tissue[J]. Nutrition & Metabolism, 2010, 7: 6. |
| [88] |
NAKAHATA Y, KALUZOVA M, GRIMALDI B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control[J]. Cell, 2008, 134(2): 329-340. DOI:10.1016/j.cell.2008.07.002 |
| [89] |
GREEN C B, TAKAHASHI J S, BASS J. The meter of metabolism[J]. Cell, 2008, 134(5): 728-742. DOI:10.1016/j.cell.2008.08.022 |
| [90] |
YAO M, HOU L G, XIE T, et al. The biosynthesis of DHA is increased in the liver of diabetic rats induced by high-fat diets and STZ, in correlation with increased activity of peroxisomal β-oxidation[J]. European Journal of Lipid Science and Technology, 2016, 118(2): 137-146. DOI:10.1002/ejlt.201400606 |
| [91] |
AHERN S.The role of the clock in lipid metabolism[D].Ph.D.Thesis.Manchester: University of Manchester, 2016.
|
| [92] |
ECKEL-MAHAN K L, PATEL V R, DE MATEO S, et al. Reprogramming of the circadian clock by nutritional challenge[J]. Cell, 2013, 155(7): 1464-1478. DOI:10.1016/j.cell.2013.11.034 |
| [93] |
KENTISH S J, VINCENT A D, KENNAWAY D J, et al. High-fat diet-induced obesity ablates gastric vagal afferent circadian rhythms[J]. Journal of Neuroscience, 2016, 36(11): 3199-3207. DOI:10.1523/JNEUROSCI.2710-15.2016 |



