蛋白质是动物必不可少的营养元素,饲粮蛋白质在动物体内水解为氨基酸(amino acids, AA)或小肽后被肠道吸收。其中,小肽(二肽和三肽)是动物摄取蛋白质的主要形式[1]。小肽的吸收利用在动物营养物质代谢和动物健康方面有重要意义。肠道小肽的吸收主要依赖小肽转运载体1(oligopeptide transporter 1, PepT1)。PepT1是一种低亲和力、高容量的膜转运蛋白,属于质子偶联寡肽转运体(proton-coupled oligopeptide transporter, POT)家族,在质子电化学梯度的驱动下,能跨膜转运寡肽和肽类药物[2-4]。PepT1主要在小肠中表达,此外,在大肠[5-6]、瓣胃和瘤胃[7]、肾脏和肝脏[5, 8]、胰腺[9]、性腺[5, 10]中亦有表达。PepT1可以转运数千种寡肽和多种肽结构类似药物,如氨基头孢菌素、血管紧张素转化酶抑制剂以及抗病毒和抗肿瘤药物[11-12]。PepT1的表达和功能已在人类[13-14]、兔[8, 15]、大鼠[16]、小鼠[17]、猪[18]、鸡[19]、牛[20-21]、绵羊[22]、牦牛[23]、斑马鱼[10]、大西洋鳕鱼[24]、大西洋鲑鱼[25]和双髻鲨[9]等多种动物中进行了研究。众多研究表明,PepT1在物种间高度保守,PepT1蛋白由大约708个AA残基组成,有12个跨膜区(transmembrane domains, TMD),且在TMD 9和TMD 10之间有1个大的细胞外环[13, 16-18, 21-22]。目前关于PepT1功能的研究主要集中在拟肽类药物转运以及胃肠道生理病理学等方面[2, 11-12, 26]。此外,PepT1的生物活性受底物、激素、年龄和生理条件等多种因素的调节[6, 27-28]。PepT1在动物的生长和代谢中起着重要的生理作用。然而,PepT1在驴中的表达和功能研究还未见报道。因此,本试验拟研究驴(Equus asinus)PepT1(ePepT1)基因的克隆、序列分析及组织表达,以期为进一步研究驴的营养物质转运提供理论依据。
1 材料与方法 1.1 总RNA的提取和全长cDNA克隆采集健康驴新鲜十二指肠组织,Trizol法提取总RNA,分光光度计测OD260 nm/OD280 nm值,琼脂糖凝胶电泳检测其完整性。根据PrimeScript® RT反转录试剂盒(TaKaRa)提供的方法进行反转录,Pfu高保真聚合酶(transgen biotech)特异扩增ePepT1 cDNA。PCR引物见表 1,PCR反应体系见表 2。PCR反应条件为:94 ℃ 3 min,98 ℃ 10 s,53 ℃ 15 s,72 ℃ 2.5 min (扩增35个循环),72 ℃ 5 min。PCR产物进行电泳检测,然后使用美基胶回收试剂盒(Magen)回收目的产物并纯化。胶回收产物连接克隆载体pTOPO-Blunt vector (Azbiochem),连接产物转化DH5α感受态细胞,涂布LB (Luria-Bertani)平板(氨苄,100 μg/mL),并于37 ℃培养过夜。挑取6个阳性菌落,进行PCR鉴定,并进行测序[生工生物工程(上海)股份有限公司]。
|
|
表 1 试验中所用引物 Table 1 Primers used in the experiment |
|
|
表 2 PCR反应体系 Table 2 PCR reaction system |
用ORF Finder推导AA序列(http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi);利用计算机pI/Mw工具(ExPASy Proteomics Server; https://web.expasy.org/compute_pi/)预测ePepT1蛋白的分子质量和理论等电点(theoretical isoelectric point, pI);利用在线软件(http://scratch.proteomics.ics.uci.edu/)预测ePepT1蛋白质的二级结构;利用生物序列分析(Lyngby TMHMM Server; http://www.cbs.dtu.dk/services/TMHMM-2.0/)预测ePepT1的跨膜区;利用NetNglyc和NetPhos服务器(http://www.cbs.dtu.dk/services/NetNGlyc/; http://www.cbs.dtu.dk/services/NetPhos/)预测潜在的N糖基化、蛋白激酶A(protein kinase A, PKA)和蛋白激酶C(protein kinase C, PKC)位点的AA序列;利用MEGA7构建系统进化树[29]。
1.3 实时荧光定量PCR试验选用2岁左右的健康德州驴公驴5头,屠宰后取肾脏、脾脏、心脏、肝脏、肺脏、胃、十二指肠、空肠和回肠组织,Trizol法分别提取各组织的总RNA。取1 mg RNA,用PrimeScript® RT试剂盒(TaKaRa)合成cDNA。然后用SYBR® PrimeScriptTM试剂盒(TaKaKa)在ABI 7500 (Applied Biosystems, Singapore)上测定ePepT1基因相对表达量。实时荧光定量PCR反应体系为20 μL。PCR引物序列见表 1。以甘油醛-3-磷酸脱氢酶(glyceraldehyde-3-phosphate dehydrogenase, GAPDH)为内参,采用2-△△Ct的方法[30]计算ePepT1基因相对表达量。每个样品重复3次,超纯水替代样品做阴性对照。
1.4 统计分析本研究所有数据用SAS 9.2进行分析,多组数据间采用单因素方差分析,数据用柱状图表示,误差线为标准差。当P < 0.05时认为差异显著。
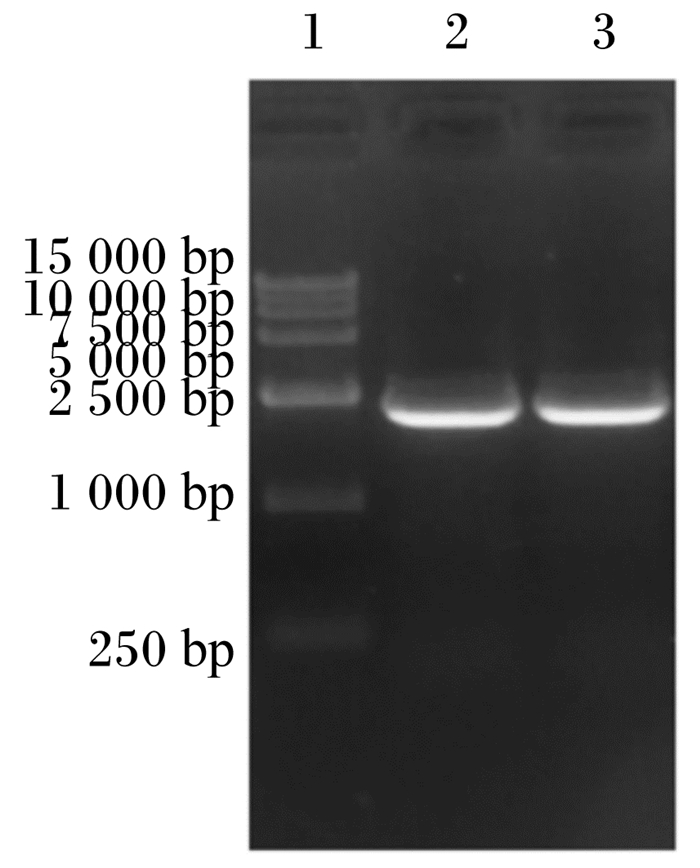
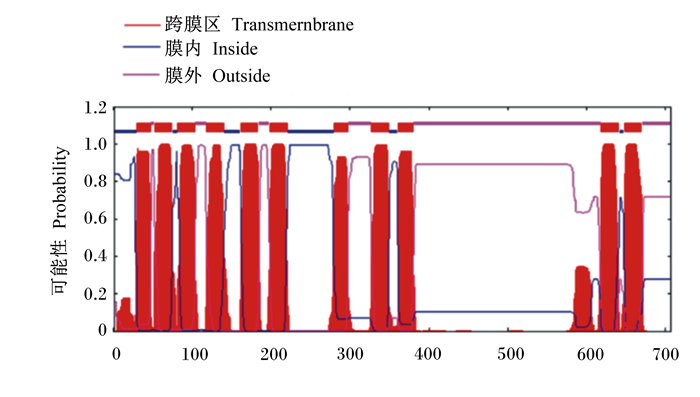
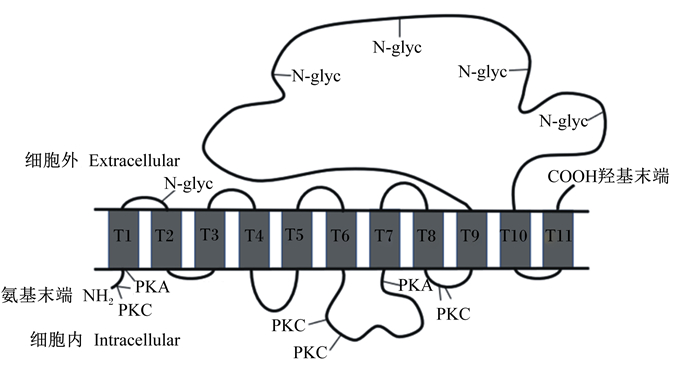
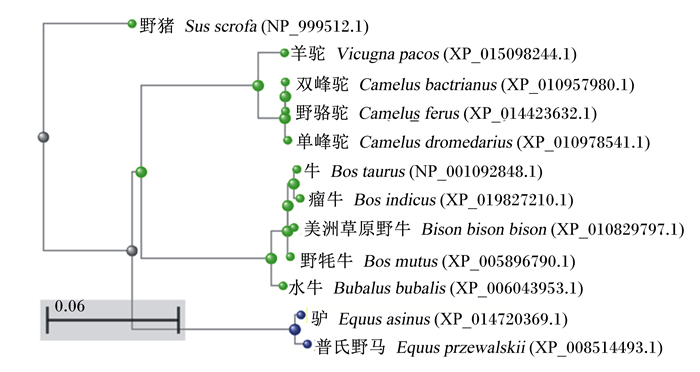
2 结果与分析 2.1 ePepT1的AA序列和系统发育分析PCR特异性扩增ePepT1基因,然后将PCR扩增产物进行电泳检测并切胶回收。目的基因的PCR产物进行琼脂糖凝胶电泳后可见一条约2 000 bp大小的条带(图 1)。经测序验证,成功扩增ePepT1基因。序列分析表明,ePepT1的开放阅读框为2 124 bp,可编码707个AA残基组成的多肽链,该蛋白分子质量为78.6 ku,等电点为7.52。ePepT1的AA序列与马(99.15%)、羊驼(86.58%)、山羊(86.28%)、牛(86.14%)、绵羊(85.43%)、猪(84.60%)、人类(83.47%)、狗(83.36%)、小鼠(83.36%)和大鼠(82.82%)的PepT1的AA序列高度保守。对ePepT1蛋白进行预测得出,二级结构中α-螺旋(Hh)占32.39%,β-折叠(Ee)占27.58%,β-转角(Tt)占9.76%,无规则卷曲(Cc)占30.27%。亲水性和疏水性分析预测ePepT1有11个潜在的TMD,且在TMD 9和TMD 10之间有1个大的细胞外环(图 2)。如图 3所示,ePepT1的AA序列中有5个胞外N-糖基化位点(Asn50、Asn438、Asn508、Asn530、Asn541),5个胞外PKC作用位点(Ser8、Ser249、Ser252、Thr356、Ser357)和3个PKA作用位点(Ser8、Ser29、Ser275)(图 3)。根据不同动物PepT1的AA序列构建的系统进化树如图 4所示。
|
1:DL15 000标记物; 2和3:ePepT1扩增产物。 1: DL15 000 marker; 2 and 3: ePepT1 amplified products. 图 1 PCR扩增产物电泳结果 Fig. 1 Electrophoresis results of PCR products |
|
横轴:氨基酸序数;纵轴:疏水系数。 Horizontal axis: amino acid number; longitudinal axis: hydrophobicity coefficient. 图 2 ePepT1蛋白的跨膜区分析 Fig. 2 Analysis of transmembrane domains of ePepT1 protein |
|
N-glyc: N糖基化位点glycosylation site;PKA: 蛋白激酶A protein kinase A; PKC:蛋白激酶C protein kinase C。 图 3 ePepT1蛋白的跨膜区模型 Fig. 3 Transmembrane domain model of ePepT1 protein |
|
图 4 基于不同动物PepT1的氨基酸序列的系统进化树 Fig. 4 Phylogenic tree constructed based on amino acid sequences of PepT1 from different animals |
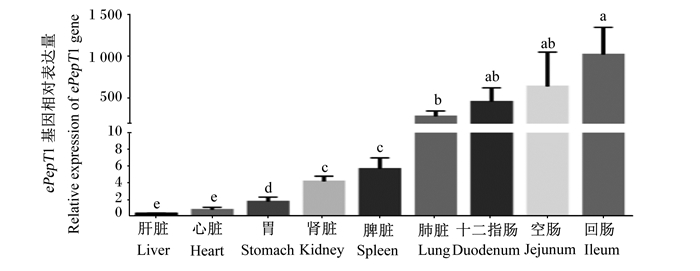
以GAPDH做内参基因,分别检测了ePepT1基因在驴肾脏、脾脏、心脏、肝脏、肺脏、胃、十二指肠、空肠、回肠组织中的表达。结果显示,ePepT1在组织中广泛表达,其在肠道(十二指肠、空肠和回肠)和肺脏中的相对表达量较高,显著高于肾脏、脾脏、胃、心脏、肝脏(P < 0.05,图 5)。
|
数据柱标注不同字母表示差异显著(P<0.05)。 Data bars with different letters mean significant difference (P < 0.05). 图 5 ePepT1基因在各组织中的相对表达量 Fig. 5 Relative expression level of ePepT1 gene in different tissues |
PepT1在从细菌到人类的不同物种中高度保守[31]。本研究发现,ePepT1 cDNA开放阅读框为2 124 bp,可以编码由707个AA残基组成、分子质量为78.6 ku的多肽链。ePepT1蛋白大小与马、绵羊和牛(707个AA残基)[21-22]相同,但比人类和猪(708个AA残基)[13, 18]、小鼠(709个AA残基)[17]以及大鼠(710个AA残基)[16]小。疏水性分析预测ePepT1有11个潜在的TMD,这与奥奈达希瓦氏菌(Shewanella oneidensis)(TMD 14)[32]和其他哺乳动物如人和牛(TMD 12)不同[13, 21]。同其他物种一样,ePepT1蛋白在TMD 9和TMD 10之间有1个大的细胞外环。此外,ePepT1蛋白TMD 9和TMD 10之间的胞外环上有5个推测的N-糖基化位点(Asn50、Asn438、Asn508、Asn530、Asn541)。N-糖基化对寡肽转运蛋白的表达、稳定性和底物转运至关重要[33-35]。Chan等[34]研究结果显示,人PepT1的TMD 9和TMD 10之间的胞外环中所有的天冬酰胺残基(asparagine, Asn)都可以被N-糖基化修饰,且N-糖基化位点的突变可以降低PepT1对甘氨酰肌氨酸(Gly-Sar)转运的Vmax值。Wang等[35]研究亦发现牛PepT2的转运功能受N-糖基化的影响。此外,ePepT1蛋白还具有5个胞外PKC作用位点(Ser8、Ser249、Ser252、Thr356、Ser357)和3个PKA作用位点(Ser8、Ser29、Ser275)。据报道,PKA和PKC可以通过调节葡萄糖转运蛋白2(GLUT2)和小肽转运蛋白PepT1来改变营养素的摄取[36]。其他研究亦证明小肽转运蛋白的转运功能受PKC磷酸化的调控[22, 37-38]。这些磷酸化位点可能在PKC和PKA调节ePepT1的转运功能中发挥重要作用。
研究发现PepT1主要在肠道中表达,其组织分布规律不同于PepT2(主要在肾脏中表达)[39]。众多研究表明,PepT1在动物消化道中广泛表达,且在十二指肠、空肠和回肠中表达量最高[5-6, 9, 24]。此外,PepT1的表达受生理状态,如发育时期、营养状况和疾病状态等多种因素的调控[6, 28, 40-41]。很多研究发现,5/6肾脏切除、小肽、甲状腺激素等均可增加PepT1的基因丰度[5, 27]。本试验中,ePepT1基因在驴肾脏、脾脏、心脏、肝脏、肺脏、胃、十二指肠、空肠、回肠组织中均有表达,且在肠道(十二指肠、空肠和回肠)中的相对表达量显著高于其他组织,其组织表达规律与以往研究结果一致。PepT1在动物体内的组织分布与其生理功能相适应,PepT1主要在消化道小肽结合氨基酸的摄取中发挥重要作用[7, 18, 21]。畜禽可通过调节PepT1的表达来改变机体对小肽的摄取,以满足不同生长阶段对营养物质的需求。本研究结果提示,ePepT1在驴肠道小肽类营养物质的吸收中发挥重要生理作用。ePepT1的表达调控及其在驴营养物质代谢中的具体作用仍需进一步研究。
4 结论① 本试验成功扩增出ePepT1基因,序列分析表明ePepT1的开放阅读框为2 124 bp,可编码707个AA残基组成的多肽链。
② 研究证实ePepT1基因在驴各组织中普遍表达,且在十二指肠、空肠和回肠中高表达。
| [1] |
ADIBI S A, MERCER D W. Protein digestion in human intestine as reflected in luminal, mucosal, and plasma amino acid concentrations after meals[J]. Journal of Clinical Investigation, 1973, 52(7): 1586-1594. DOI:10.1172/JCI107335 |
| [2] |
DANIEL H. Molecular and integrative physiology of intestinal peptide transport[J]. Annual Review of Physiology, 2004, 66: 361-384. DOI:10.1146/annurev.physiol.66.032102.144149 |
| [3] |
VERRI T, BARCA A, PISANI P, et al. Di- and tripeptide transport in vertebrates: the contribution of teleost fish models[J]. Journal of Comparative Physiology B, 2017, 187(3): 395-462. DOI:10.1007/s00360-016-1044-7 |
| [4] |
NEWSTEAD S. Recent advances in understanding proton coupled peptide transport via the POT family[J]. Current Opinion in Structural Biology, 2017, 45: 17-24. DOI:10.1016/j.sbi.2016.10.018 |
| [5] |
LU H, KLAASSEN C. Tissue distribution and thyroid hormone regulation of Pept1 and Pept2 mRNA in rodents[J]. Peptides, 2006, 27(4): 850-857. DOI:10.1016/j.peptides.2005.08.012 |
| [6] |
ZWARYCZ B, WONG E A. Expression of the peptide transporters PepT1, PepT2, and PHT1 in the embryonic and posthatch chick[J]. Poultry Science, 2013, 92(5): 1314-1321. DOI:10.3382/ps.2012-02826 |
| [7] |
PAN Y X, WONG E A, BLOOMQUIST J R, et al. Expression of a cloned ovine gastrointestinal peptide transporter (oPepT1) in Xenopus oocytes induces uptake of oligopeptides in vitro[J]. The Journal of Nutrition, 2001, 131(4): 1264-1270. DOI:10.1093/jn/131.4.1264 |
| [8] |
FEI Y J, KANAI Y, NUSSBERGER S, et al. Expression cloning of a mammalian proton-coupled oligopeptide transporter[J]. Nature, 1994, 368(6471): 563-566. DOI:10.1038/368563a0 |
| [9] |
HART H R, EVANS A N, GELSLEICHTER J, et al. Molecular identification and functional characteristics of peptide transporters in the Bonnethead shark (Sphyrna tiburo)[J]. Journal of Comparative Physiology B, 2016, 186(7): 855-866. DOI:10.1007/s00360-016-0999-8 |
| [10] |
VACCA F, BARCA A, GOMES A S, et al. The peptide transporter 1a of the zebrafish Danio rerio, an emerging model in nutrigenomics and nutrition research: molecular characterization, functional properties, and expression analysis[J]. Genes and Nutrition, 2019, 14: 33. DOI:10.1186/s12263-019-0657-3 |
| [11] |
SMITH D E, CLÉMENÇON B, HEDIGER M A. Proton-coupled oligopeptide transporter family SLC15:physiological, pharmacological and pathological implications[J]. Molecular Aspects of Medicine, 2013, 34(2/3): 323-336. |
| [12] |
OPPERMANN H, HEINRICH M, BIRKEMEYER C, et al. The proton-coupled oligopeptide transporters PEPT2, PHT1 and PHT2 mediate the uptake of carnosine in glioblastoma cells[J]. Amino Acids, 2019, 51(7): 999-1008. DOI:10.1007/s00726-019-02739-w |
| [13] |
LIANG R, FEI Y J, PRASAD P D, et al. Human intestinal H+/peptide cotransporter: cloning, functional expression, and chromosomal localization[J]. Journal of Biological Chemistry, 1995, 270(12): 6456-6463. DOI:10.1074/jbc.270.12.6456 |
| [14] |
SAITO H, MOTOHASHI H, MUKAI M, et al. Cloning and characterization of a pH-sensing regulatory factor that modulates transport activity of the human H+/peptide cotransporter, PEPT1[J]. Biochemical and Biophysical Research Communications, 1997, 237(3): 577-582. DOI:10.1006/bbrc.1997.7129 |
| [15] |
BOLL M, MARKOVICH D, WEBER W M, et al. Expression cloning of a cDNA from rabbit small intestine related to proton-coupled transport of peptides, βlactam antibiotics and ACE-inhibitors[J]. Pflügers Archiv, 1994, 429(1): 146-149. DOI:10.1007/BF02584043 |
| [16] |
SAITO H, OKUDA M, TERADA T, et al. Cloning and characterization of a rat H+/peptide cotransporter mediating absorption of beta-lactam antibiotics in the intestine and kidney[J]. Journal of Pharmacology and Experimental Therapeutics, 1995, 275(3): 1631-1637. |
| [17] |
FEI Y J, SUGAWARA M, LIU J C, et al. cDNA structure, genomic organization, and promoter analysis of the mouse intestinal peptide transporter PEPT1[J]. Biochimica et Biophysica Acta (BBA): Gene Structure and Expression, 2000, 1492(1): 145-154. DOI:10.1016/S0167-4781(00)00101-9 |
| [18] |
KLANG J E, BURNWORTH L A, PAN Y X, et al. Functional characterization of a cloned pig intestinal peptide transporter (pPepT1)[J]. Journal of Animal Science, 2005, 83(1): 172-181. DOI:10.2527/2005.831172x |
| [19] |
FRAZIER S, AJIBOYE K, OLDS A, et al. Functional characterization of the chicken peptide transporter 1 (pept1, slc15a1) gene[J]. Animal Biotechnology, 2008, 19(4): 201-210. DOI:10.1080/10495390802240206 |
| [20] |
ZIMIN A V, DELCHER A L, FLOREA L, et al. A whole-genome assembly of the domestic cow, Bos taurus[J]. Genome Biology, 2009, 10(4): R42. DOI:10.1186/gb-2009-10-4-r42 |
| [21] |
XU Q B, LIU Z X, LIU H Y, et al. Functional characterization of oligopeptide transporter 1 of dairy cows[J]. Journal of Animal Science and Biotechnology, 2018, 9(1): 7. DOI:10.1186/s40104-017-0219-8 |
| [22] |
CHEN H, PAN Y X, WONG E A, et al. Characterization and regulation of a cloned ovine gastrointestinal peptide transporter (oPepT1) expressed in a mammalian cell line[J]. The Journal of Nutrition, 2002, 132(1): 38-42. DOI:10.1093/jn/132.1.38 |
| [23] |
WANG H C, SHI F Y, HOU M J, et al. Cloning of oligopeptide transport carrier PepT1 and comparative analysis of PepT1 messenger ribonucleic acid expression in response to dietary nitrogen levels in yak (Bos grunniens) and indigenous cattle (Bos taurus) on the Qinghai-Tibetan plateau[J]. Journal of Animal Science, 2016, 94(8): 3431-3440. DOI:10.2527/jas.2016-0501 |
| [24] |
AMBERG J J, MYR C, KAMISAKA Y, et al. Expression of the oligopeptide transporter, PepT1, in larval Atlantic cod (Gadus morhua)[J]. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 2008, 150(2): 177-182. DOI:10.1016/j.cbpb.2008.02.011 |
| [25] |
GOMES A S, VACCA F, CINQUETTI R, et al. Identification and characterization of the Atlantic salmon peptide transporter 1a[J]. American Journal of Physiology: Cell Physiology, 2020, 318(1): C191-C204. DOI:10.1152/ajpcell.00360.2019 |
| [26] |
DANIEL H, KOTTRA G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology[J]. Pflügers Archiv, 2004, 447(5): 610-618. DOI:10.1007/s00424-003-1101-4 |
| [27] |
SHIMIZU Y, MASUDA S, NISHIHARA K, et al. Increased protein level of PEPT1 intestinal H+-peptide cotransporter upregulates absorption of glycylsarcosine and ceftibuten in 5/6 nephrectomized rats[J]. Gastrointestinal and Liver Physiology, 2004, 288(4): G664-G670. |
| [28] |
ALGHAMDI O A, KING N, ANDRONICOS N M, et al. Molecular changes to the rat renal cotransporters PEPT1 and PEPT2 due to ageing[J]. Molecular and Cellular Biochemistry, 2019, 452(1/2): 71-82. DOI:10.1007/s11010-018-3413-x |
| [29] |
张雨, 丰凯, 汤方. 杨小舟蛾MtroOBP1基因的克隆、序列分析及表达[J]. 南京林业大学学报(自然科学版), 2020, 44(5): 219-225. ZHANG Y, FENG K, TANG F. The cloning, sequence analysis and expression of MtroOBP1 gene in Micromelalopha troglodyte (Graeser)[J]. Journal of Nanjing Forestry University (Natural Sciences Edition), 2020, 44(5): 219-225 (in Chinese). |
| [30] |
LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR[J]. Methods, 2002, 25(4): 402-408. |
| [31] |
DANIEL H, SPANIER B, KOTTRA G, et al. From bacteria to man: archaic proton-dependent peptide transporters at work[J]. Physiology, 2006, 21(2): 93-102. DOI:10.1152/physiol.00054.2005 |
| [32] |
NEWSTEAD S. Towards a structural understanding of drug and peptide transport within the proton-dependent oligopeptide transporter (POT) family[J]. Biochemical Society Transactions, 2011, 39(5): 1353-1358. DOI:10.1042/BST0391353 |
| [33] |
JENKINS N, PAREKH R B, JAMES D C. Getting the glycosylation right: implications for the biotechnology industry[J]. Nature Biotechnology, 1996, 14(8): 975-981. DOI:10.1038/nbt0896-975 |
| [34] |
CHAN T, LU X X, SHAMS T, et al. The role of N-glycosylation in maintaining the transporter activity and expression of human oligopeptide transporter[J]. Molecular Pharmaceutics, 2016, 13(10): 3449-3456. DOI:10.1021/acs.molpharmaceut.6b00462 |
| [35] |
WANG C H, ZHAO F Q, LIU J X, et al. Short communication: the essential role of N-glycosylation in the transport activity of bovine peptide transporter 2[J]. Journal of Dairy Science, 2020, 103(7): 6679-6683. DOI:10.3168/jds.2019-16858 |
| [36] |
HIJO A H T, COUTINHO C P, ALBA-LOUREIRO T C, et al. High fat diet modulates the protein content of nutrient transporters in the small intestine of mice: possible involvement of PKA and PKC activity[J]. Heliyon, 2019, 5(10): e02611. DOI:10.1016/j.heliyon.2019.e02611 |
| [37] |
BRANDSCH M, MIYAMOTO Y, GANAPATHY V, et al. Expression and protein kinase C-dependent regulation of peptide/H+ co-transport system in the Caco-2 human colon carcinoma cell line[J]. Biochemical Journal, 1994, 299(1): 253-260. DOI:10.1042/bj2990253 |
| [38] |
WENZEL U, DIEHL D, HERGET M, et al. Regulation of the high-affinity H+/peptide cotransporter in renal LLC-PK1 cells[J]. Journal of Cellular Physiology, 1999, 178(3): 341-348. DOI:10.1002/(SICI)1097-4652(199903)178:3<341::AID-JCP8>3.0.CO;2-H |
| [39] |
DANIEL H, ZIETEK T. Taste and move: glucose and peptide transporters in the gastrointestinal tract[J]. Experimental Physiology, 2015, 100(12): 1441-1450. DOI:10.1113/EP085029 |
| [40] |
HUANG Q S, DELGADO J M V, PINOARGOTE O D S, et al. Molecular evolution of the Slc15 family and its response to waterborne copper and mercury exposure in tilapia[J]. Aquatic Toxicology, 2015, 163: 140-147. DOI:10.1016/j.aquatox.2015.04.011 |
| [41] |
卢艳娟. 日粮能量蛋白水平对滩羊主要营养素转运载体mRNA表达的影响[D]. 硕士学位论文. 杨凌: 西北农林科技大学, 2017. LU Y J. Effects of diets with different energy and protein levels on mrna expression of nutrient transporters in Tan sheep[D]. Master's Thesis. Yangling: Northwest Agriculture & Forestry University, 2007. (in Chinese) |



