2. 西藏自治区 农牧科学院畜牧兽医研究所, 拉萨 850009;
3. 四川省草原科学研究院, 成都 611731;
4. 四川省红原县刷经寺农业农村发展中心, 阿坝 624402
2. Institute of Animal Husbandry and Veterinary Medicine, Tibet Academy of Agriculture and Animal Husbandry Science, Lhasa 850009, China;
3. Sichuan Academy of Grassland Science, Chengdu 611731, China;
4. Hongyuan County, Sichuan Province Shuajingsi Center for Agricultural Rural Development, Aba 624402, China
牧区牧草季节性生长,牦牛犏牛在春冬季往往没有充足的饲草资源,常常造成牛只掉膘,甚至是死亡的发生。而肉牛的“北繁南育”是协调南北饲料资源,降低生产成本,创造经济收入的重要举措[1]。但是,由于生存环境和饲料的剧烈变化以及长途运输带来的多种综合应激往往导致牛采食量下降,免疫功能下降,发病率暴增,死亡率居高不下。因此,迅速提高长途运输后的异地育肥牛只的干物质采食量(DMI)、恢复体质是降低疾病发生率和死亡率的关键。
酵母培养物(yeast culture,YC)是酵母菌在特定的培养基中发酵后形成的微生态制品,富含蛋白质、寡糖和有机酸等成分。Dann等[2]研究表明,在围产期奶牛饲粮中添加YC可提高其DMI和泌乳早期产奶量,减少体储消耗。Wagner等[3]的研究发现,YC可以显著提高育肥牛的DMI与平均日增重(ADG)。糖蜜(molasses,MO)是制糖业副产品,主要成分是糖。Havekes等[4]报道,MO具有显著提高干奶期奶牛DMI,改善瘤胃健康的作用。Mordent等[5]报道,MO可以提高反刍动物的DMI,并且对提高放牧肉牛、水牛、绵羊和山羊的DMI效果更佳。前人研究大都从YC和MO本身如口感和香味等角度出发去探究其提高动物DMI的原因,本试验拟考察YC和MO对异地育肥肉牛的影响,并尝试从微生物的角度去解释YC和MO改变肉牛DMI的原因。
1 材料与方法 1.1 试验材料YC购自某生物发酵工程技术(深圳)有限公司。MO是甘蔗糖蜜,购自当地工厂。
1.2 试验动物与设计选取32头体重[(107.0±14.7) kg]和日龄相近的肉牛生长牛(牦牛♂×犏牛♀)作为试验动物,随机分为4个组,每组8头牛。对照组(CK组)饲喂基础饲粮,3个试验组分别饲喂基础饲粮+YC(30 g/d)(YC组)、MO饲粮(MO组)和MO饲粮+YC(30 g/d)(YM组)。整个试验周期70 d。
1.3 饲粮组成及营养水平饲粮配方设计参照中国《肉牛饲养标准》(NY/T 815—2004),基础饲粮与MO饲粮的营养水平相近,除了添加MO以外,饲粮结构保持不变。饲粮组成及营养水平见表 1。
|
|
表 1 饲粮组成及营养水平(风干基础) Table 1 Composition and nutrient levels of diets (air-dry basis) |
试验于2020年10—12月在四川省成都市都江堰市某牛场完成。试验开始前对牛圈进行清洁和消毒等准备工作。试验牛只进行舍饲分栏栓养。每天07:30和18:30,以全混合日粮(TMR)的形式饲喂,自由采食与饮水。
1.5 样品采集及指标测定 1.5.1 生长性能于试验第1和70天晨饲前对试验牛称重,记录试验牛每天的采食量。计算ADG、DMI、料重比(F/G)。
1.5.2 营养物质表观消化率于试验第66~69天每4 h采集1次粪样,采集完成后将每头试验牛的粪样混合均匀加入10%的稀硫酸固氮,在此期间收集饲料样品。粪样和饲料样65 ℃烘至恒重,回潮24 h后粉碎过40目筛保存。
粪样和饲料样中干物质(DM)、中性洗涤纤维(NDF)、酸性洗涤纤维(ADF)和粗蛋白质(CP)含量分别参照GB/T 6435—2014、GB/T 20806—2006、NY/T 1459—2007和GB/T 6432—2018测定。营养物质表观消化率采用盐酸不溶灰分(AIA)法测定,计算公式如下:

|
于试验第69天晨饲前,用无菌棉签刮取试验牛直肠末端粪样约5 g,冻存管液氮保存到实验室后转移至-80 ℃冰箱保存待测。
使用Zymo Research BIOMICS DNA Microprep Kit(Cat# D4301)进行样本DNA提取与纯化;使用Primer5′-3′对样本16S rDNA V4区域进行PCR扩增,通用引物为515F(5′-GTGYCAGCMGCCGCGGTAA-3′)和806R(5′-GGACTACHVGGGTWTCTAAT-3′);2%凝胶电泳检测,对合格的样品使用Zymoclean Gel Recovery Kit(D4008)回收;使用Qubit@2.0 Fluorometer (Thermo Scientific)定量;在Illumina Hiseq 2500平台进行测序。
使用FLASH拼接双端序列得到原始Tags(raw tags)使用QIIME进行质控得到高质量Tags(clean tags),使用UCHIME算法与数据库比对去除嵌合体得到有效Tags(effective Tags),使用Usearch在97%水平上进行操作分类单元(OTU)聚类,使用UCLUST分类法与SILVA数据库进行物种分类注释。
1.6 统计分析试验数据经Excel 2016整理后,用SPSS 27.0对生长性能和营养物质表观消化率数据进行单因素方差分析(one-way ANOVA),并以Duncan氏法进行多重比较;对微生物数据进行非参数检验(Kruskal-Wallis)。P < 0.05表示差异显著,P < 0.01表示差异极显著。
2 结果 2.1 添加YC和MO对肉牛生长性能的影响由表 2可知,3个试验组的DMI均显著高于CK组(P < 0.05),分别提高了7.80%、9.57%和14.89%,但3个试验组之间差异不显著(P>0.05);4个组之间的初始体重(IBW)、终末体重(FBW)、ADG和F/G差异不显著(P>0.05)。
|
|
表 2 YC和MO对肉牛生长性能的影响 Table 2 Effects of YC and MO on growth performance of beef cattle |
由表 3可知,YC组和YM组NDF表观消化率极显著高于CK组(P < 0.01),YM组显著高于CK组(P < 0.05),3个试验组ADF表观消化率显著高于CK组(P < 0.05),但3个试验组之间差异不显著(P>0.05)。4个组之间的DM和CP表观消化率差异不显著(P>0.05)。
|
|
表 3 YC和MO对肉牛营养物质表观消化率的影响 Table 3 Effects of YC and MO on nutrient apparent digestibility of beef cattle |
由表 4可知,YM组的OTU极显著高于CK组(P < 0.01),显著高于YC组(P < 0.05),MO组的OTU显著高于CK组(P < 0.05)。YM组的Chao1指数和ACE指数显著高于CK组、YC组和MO组(P < 0.05),CK组、YC组和MO组之间的Chao1指数和ACE指数差异不显著(P>0.05);3个试验组的Shannon指数和Simpson指数极显著高于CK组(P < 0.01),但3个试验组之间差异不显著(P>0.05)。
|
|
表 4 粪便微生物Alpha多样性 Table 4 Alpha diversity of fecal microorganism |
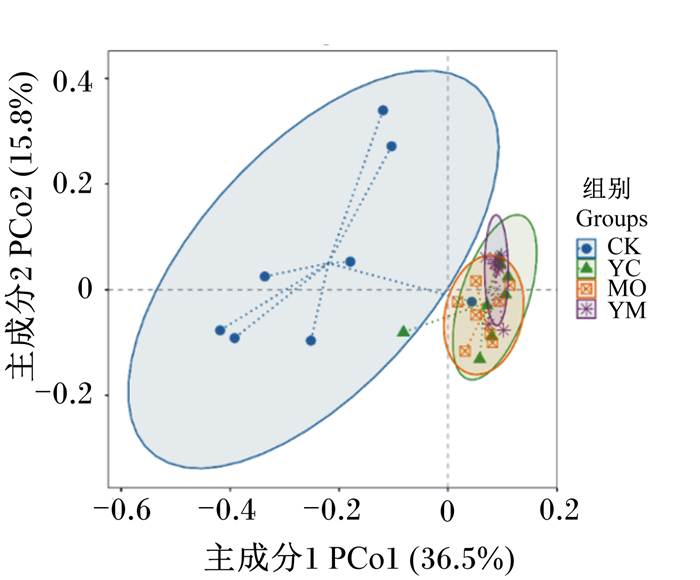
由图 1可知,主坐标1(PCo1)的贡献值为36.5%,主坐标2(PCo2)的贡献值为15.8%,3个试验组跟CK组明显区分开,而3个试验组交叉在一起。CK组的8个样品较为分散,而3个试验组的样品间距离较近,均一性较好。
|
图 1 粪便微生物群落构成的主坐标分析图 Fig. 1 PCoA diagram of fecal microorganism composition |
本试验在生长肉牛的粪便中共鉴定出微生物35个门(phylum)、77个纲(class)、163个目(order)、287个科(family)和644个属(genus)。
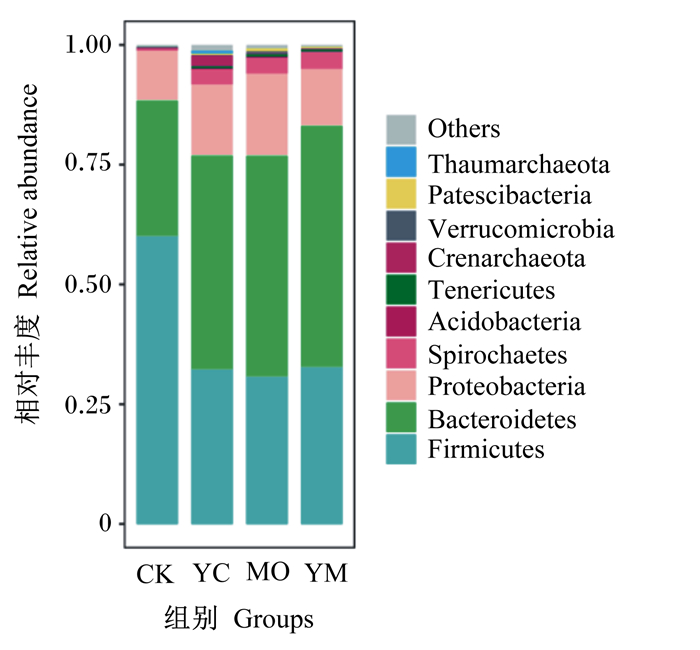
相对丰度排名前10的菌门如图 2所示,拟杆菌门(Bacteroidetes)、厚壁菌门(Firmicutes)和变形菌门(Proteobacteria)3个菌门是粪便优势菌门,占总细菌相对丰度91.73%以上。由表 5可知,3个试验组的拟杆菌门的相对丰度显著高于CK组(P < 0.05),螺旋体门(Spirochaetes)相对丰度极显著高于CK组(P < 0.01),但3个试验组之间差异不显著(P>0.05)。MO组和YM组软壁菌门(Tenericutes)和髌骨细菌门(Patescibacteria)的相对丰度极显著高于CK组(P < 0.01), YC组显著高于CK组(P < 0.05)。MO组和YM组厚壁菌门的相对丰度极显著低于CK组(P < 0.01),YC组显著低于CK组(P < 0.05)。4个组之间的变形菌门、酸杆菌门(Acidobacteria)、泉古菌门(Crenarchaeota)、疣微菌门(Verrucomicrobia)、奇古菌门(Thaumarchaeota)的相对丰度差异不显著(P>0.05)。
|
Firmicutes:厚壁菌门;Bacteroidetes:拟杆菌门;Proteobacteria:变形菌门;Spirochaetes:螺旋体门;Acidobacteria:酸杆菌门;Tenericutes:软壁菌门;Crenarchaeota:泉古菌门;Verrucomicrobia:疣微菌门;Patescibacteria:髌骨细菌门;Thaumarchaeota:奇古菌门;Others:其他。 图 2 门水平上粪便微生物群落相对丰度 Fig. 2 Relative abundance of fecal microbial community at phylum level |
|
|
表 5 粪便微生物群落在门水平上的分布 Table 5 Distribution of fecal microbial community at phylum level |
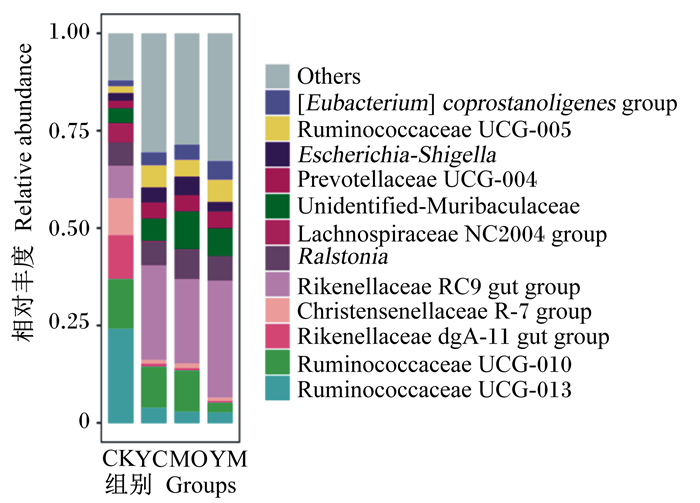
12种优势菌属(相对丰度>1%)如图 3所示。由表 6可知,YC组和YM组理研菌科RC9肠道群(Rikenellaceae RC9 gut group)和瘤胃球菌科UCG-005(Ruminococcaceae UCG-005)相对丰度极显著高于CK组(P < 0.01),MO组显著高于CK组(P < 0.05)。MO组和YM组鼠杆菌科未定义菌属(Unidentified-Muribaculaceae)和类杆菌真核菌群([Eubacterium] coprostanoligenes group)相对丰度极显著高于CK组(P < 0.01),YC组显著高于CK组(P < 0.05)。3个试验组的普雷沃氏菌科UCG-004(Prevotellaceae UCG-004)相对丰度显著高于CK组(P < 0.05)。YC组和YM组克里斯滕森氏菌科R-7群(Christensenellaceae R-7 group)相对丰度极显著低于CK组(P < 0.01),MO组显著低于CK组(P < 0.05)。4个组之间的瘤胃球菌科UCG-013(Ruminococcaceae UCG-013)、瘤胃球菌科UCG-010(Ruminococcaceae UCG-010)、理研菌科dgA-11肠道群(Rikenellaceae dgA-11 gut group)、青枯菌属(Ralstonia)、毛螺菌科NC2004(Lachnospiraceae NC2004 group)和大肠埃希菌-志贺菌属(Escherichia-Shigella)的相对丰度无显著差异(P>0.05)。
|
Ruminococcaceae UCG-013:瘤胃球菌科UCG-013;Ruminococcaceae UCG-010:瘤胃球菌科UCG-010;Rikenellaceae dgA-11 gut group:理研菌科dgA-11肠道群;Christensenellaceae R-7 group:克里斯滕森氏菌科R-7;Rikenellaceae RC9 gut group:理研菌科RC9肠道群;Ralstonia:青枯菌属;Lachnospiraceae NC2004 group:毛螺菌科NC2004群;Unidentified-Muribaculaceae:鼠杆菌科未定义属;Prevotellaceae UCG-004:普雷沃氏菌科UCG-004;Escherichia-Shigella:大肠埃希菌-志贺菌属;Ruminococcaceae UCG-005:瘤胃球菌科UCG-005;[Eubacterium] coprostanoligenes group:类杆菌真核菌群;Others:其他。 图 3 属水平上粪便微生物群落相对丰度 Fig. 3 Relative abundance of fecal microbial community at genus level |
|
|
表 6 粪便微生物群落在属水平上的分布 Table 6 Distribution of fecal microbial community at genus level |
异地育肥是我国肉牛生产的主要方式,但由于长途运输中的缺水缺料以及新气候环境等多种应激,常常导致牛只DMI的下降,久久不能恢复,而DMI则直接影响动物机体健康和生产力[6],DMI的恢复对动物机体免疫力恢复与减少疾病发生有重要意义。本试验在饲粮中添加YC肉牛DMI显著提高,与前人研究结果一致。Lesmeister等[7]在饲粮中添加YC发现犊牛DMI显著提高。黄文明等[8]认为YC能提高牛只的DMI的原因是其芳香气味提高了牛只的食欲。本试验在饲粮中添加MO的肉牛DMI显著提高,前人也有相似的报道。Børsting等[9]研究表明,在奶牛饲粮中添加MO可以显著提高泌乳期奶牛的DMI。Devries等[10]认为MO能提高反刍动物DMI是因为MO可以改善饲粮适口性,大多数动物喜食甜食。但YM组与YC组和MO组相比DMI差异不显著,这表明同时添加YC和MO对肉牛DMI没有进一步的叠加效应。
3.2 添加YC和MO对肉牛营养物质表观消化率的影响饲粮中营养物质表观消化率不仅是动物消化能力的直接反映,也是影响动物生长性能的重要因素。Robinson[11]研究表明,YC可以有效提高产后奶牛的NDF、ADF等营养物质表观消化率。本试验在肉牛饲粮中添加YC也观察到NDF和ADF表观消化率显著提高。Haddad等[12]在育肥羔羊中也发现,添加YC可以显著提高育肥羔羊的NDF和ADF表观消化率。Mordenti等[5]报道,MO对提高反刍动物营养物质表观消化率有重要作用,本试验在肉牛饲粮中添加MO发现NDF和ADF表观消化率显著提高,但Zali等[13]报道,MO显著提高了荷斯坦公犊牛的NDF表观消化率,但对于ADF表观消化率的影响不显著,这可能是因为牛的品种和饲粮结构不同所致。混合添加YC和MO发现NDF和ADF表观消化率显著高于CK组,但3个试验组之间的NDF和ADF表观消化率差异不显著,这表明同时添加YC和MO对提高营养物质的表观消化率无叠加效应,这也从不同处理组牛只的生长性能上体现了出来。纤维类物质进入胃之后与大量水结合膨大形成凝胶状团状物,增大食糜黏度,使胃肠道蠕动减缓,导致动物饱腹感增强,从而降低动物采食欲望[14]。研究表明纤维类物质消化率提高有助于泌乳奶牛DMI的提高[15]。3个试验组的NDF和ADF表观消化率均高于CK组,这可能是3个试验组DMI高于CK组的原因。
3.3 添加YC和MO对肉牛粪便微生物区系组成的影响饲粮中纤维类物质的消化率提高或许跟肠道微生物区系组成的改变有关。反刍动物胃肠道微生物对其摄入的营养物质降解有重要作用[16]。纤维类物质的降解主要依靠微生物降解,而瘤胃是纤维类物质降解的主要场所[17],但是研究表明反刍动物后肠段对纤维物质的降解也具有重要作用,约30%的纤维类物质需要依靠后肠微生物来降解[18]。而粪便微生物可以用来评估反刍动物后肠段的微生物区系组成[19]。在本试验中发现,肉牛的粪便微生物优势菌门分别是厚壁菌门、拟杆菌门和变形菌门,这与前人在野牛[19]、犊牛[20]以及山羊[21]上的报道结果一致。本试验在饲粮中添加YC、MO或二者同时添加均提高了拟杆菌门的相对丰度,并降低了厚壁菌门的相对丰度。研究表明,拟杆菌门是健康人类粪便中相对丰度最高的菌群[22],可以帮助宿主高效的吸收与利用多糖[23]。而厚壁菌门的作用主要是协助多糖发酵,和拟杆菌门一起帮助宿主消化一些自身无法利用的物质[24]。饲粮中添加YC、MO或两者混合添加可以提高拟杆菌门相对丰度,这可能有助于提高肉牛的纤维降解消化能力。
本试验在饲粮中添加YC、MO或二者混合添加均提高了理研菌科RC9肠道群、鼠杆菌科未定义菌属、普雷沃氏菌科UCG-004、瘤胃球菌科UCG-005和类杆菌真核菌群的相对丰度。Zhu等[25]研究表明,理研菌科RC9肠道群可以有效降解可溶性多糖和不溶性纤维素,这或许是在饲粮中添加YC、MO或二者同时添加肉牛NDF与ADF表观消化率显著提高的原因之一。也有研究表明,理研菌科菌属对维护肠道健康有重要作用[26]。Li等[27]报道,普雷沃氏菌属可以降解淀粉、蛋白质、纤维素和半纤维素等。也有研究表明普雷沃氏菌属有协作降解纤维的能力[28],这可能也是本试验中3个试验组的NDF与ADF表格消化率显著高于CK组的原因。此外,有研究表明普雷沃氏菌属对降低肠道炎症方面也具有重要作用[29]。瘤胃球菌科UCG-005和类杆菌真核菌群均属于瘤胃球菌科,前人的研究表明,瘤胃球菌科菌属是重要的纤维降解菌,可以降解宿主不能消化的纤维类物质[30]。Patra等[31]和Zhao等[32]的研究表明,瘤胃中瘤胃球菌科菌属的相对丰度跟饲粮的纤维消化率呈正相关,这也是本试验中3个试验组的NDF与ADF表观消化率显著高于CK组的原因。本试验中3个试验组的克里斯滕森氏菌科R-7的相对丰度低于CK组,而前人的报道表明,克里斯滕森氏菌属的相对丰度与牦牛增重呈负相关[33]。本试验中3个试验组与CK组相比粪便微生物发生了相似的变化,且这些变化的菌群均对动物的纤维物质的消化、肠道代谢与健康有一定的积极作用,从而使NDF和ADF表观消化率提高,进而提高DMI。
4 结论添加YC、MO或两者混合添加均可以提高肉牛胃肠道中拟杆菌门的相对丰度,提高理研菌科RC9肠道群和普雷沃氏菌科UCG-004等菌属的相对丰度,从而提高NDF和ADF表观消化率,进而提高DMI,但两者混合添加对改善肉牛消化率和采食量并无叠加效应。
| [1] |
张娇娇. 饲用缓释尿素对后备奶牛和肉牛体外瘤胃发酵参数、生长性能和血液指标的影响[D]. 硕士学位论文. 兰州: 兰州大学, 2018. ZHANG J J. Effects of feeding slow-release urea on in vitro rumen fermentation parameters, growth performance and blood indexes of reserve dairy cows and beef cattle[D]. Master's Thesis. Lanzhou: Lanzhou University, 2018. (in Chinese) |
| [2] |
DANN H M, DRACKLEY J K, MCCOY G C, et al. Effects of yeast culture (Saccharomyces cerevisiae) on prepartum intake and postpartum intake and milk production of Jersey cows[J]. Journal of Dairy Science, 2000, 83(1): 123-127. DOI:10.3168/jds.S0022-0302(00)74863-6 |
| [3] |
WAGNER J J, ENGLE T E, BELKNAP C R, et al. Meta-analysis examining the effects of Saccharomyces cerevisiae fermentation products on feedlot performance and carcass traits1, 2, 3[J]. The Professional Animal Scientist, 2016, 32(2): 172-182. DOI:10.15232/pas.2015-01438 |
| [4] |
HAVEKES C D, DUFFIELD T F, CARPENTER A J, et al. Effects of molasses-based liquid feed supplementation to a high-straw dry cow diet on feed intake, health, and performance of dairy cows across the transition period[J]. Journal of Dairy Science, 2020, 103(6): 5070-5089. DOI:10.3168/jds.2019-18085 |
| [5] |
MORDENTI A L, GIARETTA E, CAMPIDONICO L, et al. A review regarding the use of molasses in animal nutrition[J]. Animals, 2021, 11(1): 115. DOI:10.3390/ani11010115 |
| [6] |
EASTRIDGE M L. Major advances in applied dairy cattle nutrition[J]. Journal of Dairy Science, 2006, 89(4): 1311-1323. DOI:10.3168/jds.S0022-0302(06)72199-3 |
| [7] |
LESMEISTER K E, HEINRICHS A J, GABLER M T. Effects of supplemental yeast (Saccharomyces cerevisiae) culture on rumen development, growth characteristics, and blood parameters in neonatal dairy calves[J]. Journal of Dairy Science, 2004, 87(6): 1832-1839. DOI:10.3168/jds.S0022-0302(04)73340-8 |
| [8] |
黄文明, 谭林, 王芬, 等. 酵母培养物对育肥牛生长性能、屠宰性能及肉品质的影响[J]. 动物营养学报, 2019, 31(3): 1317-1325. HUANG W M, TAN L, WANG F, et al. Effects of yeast culture on growth performance, slaughter performance and meat quality of finishing cattle[J]. Chinese Journal of Animal Nutrition, 2019, 31(3): 1317-1325 (in Chinese). |
| [9] |
BØRSTING C F, BRASK M, HELLWING A L F, et al. Enteric methane emission and digestion in dairy cows fed wheat or molasses[J]. Journal of Dairy Science, 2020, 103(2): 1448-1462. DOI:10.3168/jds.2019-16655 |
| [10] |
DEVRIES T J, GILL R M. Adding liquid feed to a total mixed ration reduces feed sorting behavior and improves productivity of lactating dairy cows[J]. Journal of Dairy Science, 2012, 95(5): 2648-2655. DOI:10.3168/jds.2011-4965 |
| [11] |
ROBINSON P H. Effect of yeast culture (Saccharomyces cerevisiae) on adaptation of cows to diets postpartum[J]. Journal of Dairy Science, 1997, 80(6): 1119-1125. DOI:10.3168/jds.S0022-0302(97)76038-7 |
| [12] |
HADDAD S G, GOUSSOUS S N. Effect of yeast culture supplementation on nutrient intake, digestibility and growth performance of Awassi lambs[J]. Animal Feed Science and Technology, 2005, 118(3/4): 343-348. |
| [13] |
ZALI A, EFTEKHARI M, FATEHI F, et al. Effect of vinasse (condensed molasses solubles) on performance and meat chemical composition of Holstein male calves[J]. Italian Journal of Animal Science, 2017, 16(3): 515-520. DOI:10.1080/1828051X.2017.1298407 |
| [14] |
STEINGOETTER A, BUETIKOFER S, CURCIC J, et al. The dynamics of gastric emptying and self-reported feelings of satiation are better predictors than gastrointestinal hormones of the effects of lipid emulsion structure on fat digestion in healthy adults-a Bayesian inference approach[J]. The Journal of Nutrition, 2017, 147(4): 706-714. DOI:10.3945/jn.116.237800 |
| [15] |
CHEN B, WANG C, WANG Y M, et al. Effect of biotin on milk performance of dairy cattle: a Meta-analysis[J]. Journal of Dairy Science, 2011, 94(7): 3537-3546. DOI:10.3168/jds.2010-3764 |
| [16] |
MALMUTHUGE N, GUAN L L. Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation[J]. Journal of Animal Science and Biotechnology, 2017, 8(1): 8. DOI:10.1186/s40104-016-0135-3 |
| [17] |
马健, MUJTABA S A, 王之盛. 瘤胃微生物区系的影响因素及其调控措施[J]. 动物营养学报, 2020, 32(5): 1957-1964. MA J, MUJTABA S A, WANG Z S. Influencing factors and regulating measures of rumen microbial flora[J]. Chinese Journal of Animal Nutrition, 2020, 32(5): 1957-1964 (in Chinese). DOI:10.3969/j.issn.1006-267x.2020.05.001 |
| [18] |
HOOVER W H. Digestion and absorption in the hindgut of ruminants[J]. Journal of Animal Science, 1978, 46(6): 1789-1799. DOI:10.2527/jas1978.4661789x |
| [19] |
DE OLIVEIRA M N V, JEWELL K A, FREITAS F S, et al. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer[J]. Veterinary Microbiology, 2013, 164(3/4): 307-314. |
| [20] |
OIKONOMOU G, TEIXEIRA A G V, FODITSCH C, et al. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth[J]. PloS One, 2013, 8(4): e63157. DOI:10.1371/journal.pone.0063157 |
| [21] |
HUO W, ZHU W, MAO S. Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats[J]. World Journal of Microbiology and Biotechnology, 2014, 30(2): 669-680. DOI:10.1007/s11274-013-1489-8 |
| [22] |
ZITOMERSKY N L, COYNE M J, COMSTOCK L E. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut[J]. Infection and Immunity, 2011, 79(5): 2012-2020. DOI:10.1128/IAI.01348-10 |
| [23] |
MURPHY E F, COTTER P D, HEALY S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models[J]. Gut, 2010, 59(12): 1635-1642. DOI:10.1136/gut.2010.215665 |
| [24] |
RUIZ L, MARGOLLES A, SÁNCHEZ B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium[J]. Frontiers in Microbiology, 2013, 4: 396. |
| [25] |
ZHU Y X, WANG Z S, HU R, et al. Comparative study of the bacterial communities throughout the gastrointestinal tract in two beef cattle breeds[J]. Applied Microbiology and Biotechnology, 2021, 105(1): 313-325. DOI:10.1007/s00253-020-11019-7 |
| [26] |
COX L M, YAMANISHI S, SOHN J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences[J]. Cell, 2014, 158(4): 705-721. DOI:10.1016/j.cell.2014.05.052 |
| [27] |
LI F Y, GUAN L L. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle[J]. Applied and Environmental Microbiology, 2017, 83(9): e00061-17. |
| [28] |
LI F, GUAN L L. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle[J]. Applied & Environmental Microbiology, 2017, 83(9): 1-17. |
| [29] |
FORSYTH C B, SHANNON K M, KORDOWER J H, et al. Increased inte stinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson's disease[J]. Plos One, 2011, 6(12): e28032. DOI:10.1371/journal.pone.0028032 |
| [30] |
BIDDLE A, STEWART L, BLANCHARD J, et al. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities[J]. Diversity, 2013, 5(3): 627-640. DOI:10.3390/d5030627 |
| [31] |
PATRA A K, YU Z T. Essential oils affect populations of some rumen bacteria in vitro as revealed by microarray (Rumen Bact Array) analysis[J]. Frontiers in Microbiology, 2015, 6: 297. |
| [32] |
ZHAO X H, CHEN Z D, ZHOU S, et al. Effects of daidzein on performance, serum metabolites, nutrient digestibility, and fecal bacterial community in bull calves[J]. Animal Feed Science and Technology, 2017, 225: 87-96. DOI:10.1016/j.anifeedsci.2017.01.014 |
| [33] |
MA J, ZHU Y X, WANG Z S, et al. Comparing the bacterial community in the gastrointestinal tracts between growth-retarded and normal yaks on the Qinghai-Tibetan plateau[J]. Frontiers in Microbiology, 2020, 11: 600516. DOI:10.3389/fmicb.2020.600516 |




