2. 四川农业大学动物营养研究所, 教育部动物抗病营养重点实验室, 成都 611100
2. Key laboratory for Animal Disease Resistance Nutrition of Ministry of Education, Animal Nutrition Institute, Sichuan Agricultural University, Chengdu 610000, China
胆汁酸是由胆固醇合成的具有两性性质(亲水性和疏水性)的分子。近年来发现,胆汁酸除了通过形成混合胶束促进肠道对脂类物质消化吸收之外,还作为信号分子通过胆汁酸受体感知营养,调控胆汁酸和胆固醇稳态、糖脂代谢、免疫反应以及介导宿主与肠道微生物交互作用等[1]。胆盐水解酶(bile salt hydrolase,BSH)介导结合型胆汁酸(conjugated bile acid,CBA)转化为非结合型胆汁酸(unconjugated bile acid,UCBA)和甘氨酸或牛磺酸,是肠道微生物介导初级胆汁酸转化为次级胆汁酸的关键步骤。近期越来越多研究发现,BSH对肠道微生物和宿主生理代谢具有重要调节作用。一方面,BSH有助于微生物在肠道定植和黏附[2];另一方面,BSH通过调节胆汁酸代谢,进而影响宿主对饲粮脂类消化吸收、胆固醇代谢、能量代谢和炎症反应[3-4]。本文主要围绕BSH结构、分布、生物学功能以及BSH抑制剂促进动物生长作用及其研究进展等方面进行简要综述,以期为全面了解BSH功能,并对其在畜牧业中应用提供参考。
1 BSH简介 1.1 BSH结构BSH是一类N-末端亲核胞内水解酶,介导胆汁中结合胆盐生成游离胆汁酸和甘氨酸或牛磺酸。BSH通常含有N端半胱氨酸残基,在起始甲酰甲硫氨酸蛋白水解后,N端半胱氨酸残基成为其催化中心。半胱氨酸残基中SH基团对于BSH活性起着至关重要作用。研究发现,若将BSH半胱氨酸残基替换为丝氨酸、苏氨酸或丙氨酸等不具有SH基团的氨基酸时,其酶活性均会消失[5];采用氧化剂[麦古利苯甲酸、碘乙酰胺、汞离子(Hg2+)、铜离子(Cu2+)和镉离子(Cd2+)]氧化SH基团也会导致BSH活性受到抑制[5-7]。此外,天冬氨酸(Asp)-20、酪氨酸(Tyr)-82、天冬酰胺(Asn)-175和精氨酸(Arg)-228通常也被认为在BSH活性中发挥着关键作用。对两歧双歧杆菌、长双歧杆菌、产气荚膜梭菌、嗜酸乳杆菌、约氏乳酸杆菌、植物乳酸杆菌、单核细胞增多性李斯特氏菌和环形芽孢杆菌BSH蛋白序列比对发现,半胱氨酸、天冬氨酸、天冬酰胺和精氨酸在BSH中均高度保守[8]。
1.2 BSH与胆汁酸代谢胆汁酸主要由肝脏实质细胞负责合成,我们通常称肝脏胆汁酸合成通路中的直接产物为初级胆汁酸。初级胆汁酸合成主要包括胆固醇类固醇环C7或支链C24、C25和C27羟基化,类固醇环修饰和支链缩短[9]。胆汁酸在体内代谢主要通过肠肝循环。在胆汁酸分泌进入胆管腔之前,初级胆汁酸24号碳原子在胆汁酰辅酶A合成酶(bile acid-CoA synthetase,BACS)和胆汁酸辅酶A: 氨基酸N-酰基转移酶(bile acid coenzyme A: amino acid N-acyltransferase,BAAT)作用下与甘氨酸或牛磺酸发生结合产生CBA,包括甘氨胆酸(glycine-conjugated bile acids,G-CBA)或牛磺胆酸(taurine-conjugated bile acids,T-CBA)[10]。鉴于BAAT具有极高活性,超过98%的肝脏胆汁酸在分泌之前发生结合反应。BAAT催化甘氨酸或牛磺酸与初级胆汁酸结合反应与物种密切相关,人和猪上CBA主要为G-CBA,而啮齿动物以T-CBA为主[11-12]。
肠道是次级胆汁酸的主要合成场所,肠道微生物是其执行者。肠道微生物对胆汁酸代谢包括去结合反应,C-3、C-7和C-12位羟基氧化以及7α/β-脱羟基作用[13]。目前已知次级胆汁酸超过20种,且存在物种差异(图 1)。需要指出,猪的次级胆汁酸包括猪脱氧胆酸(hyodeoxycholic acid,HDCA)、脱氧胆酸(deoxycholic acid,DCA)、石胆酸(lithocholic acid,LCA)和熊脱氧胆酸等[10]。BSH介导的去结合反应是肠道微生物参与次级胆汁酸合成的关键步骤[14],详细过程见图 1。无菌小鼠和抗生素处理均会导致肠腔中CBA大量累积,次级胆汁酸水平急剧降低[15]。此外,无菌小鼠定植BSH敲除多形拟杆菌显著影响宿主胆汁酸代谢功能[16]。
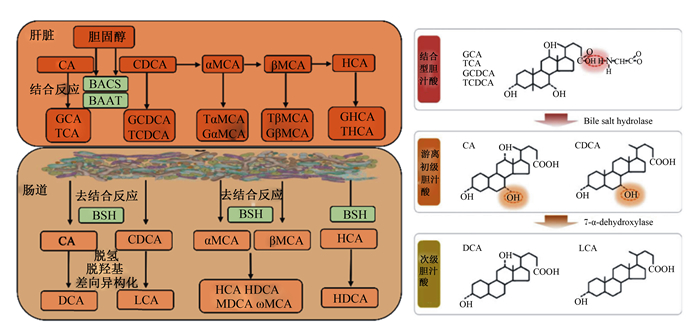
|
BACS:胆汁酸辅酶A合成酶bile acid-CoA synthetase;BAAT:胆汁酸辅酶A氨基酸N-乙酰转移酶bile acid-CoA: amino acid N-acetyltransferase;BSH:胆盐水解酶bile salt hydrolase;CA:胆酸cholic acid;GCA:甘氨胆酸glycine-conjugated cholic acid;TCA:牛磺胆酸taurine-conjugated cholic acid;CDCA:鹅脱氧胆酸chenodeoxycholic acid;GCDCA:甘氨鹅脱氧胆酸glycine-conjugated chenodeoxycholic acid;TCDCA:牛磺鹅脱氧胆酸taurine-conjugated chenodeoxycholic acid;DCA:脱氧胆酸deoxycholic acid;LCA:石胆酸lithocholic acid;α-MCA:α-鼠胆酸alpha muricholic acid;β-MCA:β-鼠胆酸beta muricholic acid;Gα-MCA:甘氨α-鼠胆酸glycine-conjugated alpha muricholic acid;Tα-MCA:牛磺α-鼠胆酸taurine-conjugated alpha muricholic acid;Gβ-MCA:甘氨β-鼠胆酸glycine-conjugated beta muricholic acid;Tβ-MCA:牛磺β-鼠胆酸taurine-conjugated beta muricholic acid;HCA:猪胆酸hyocholic acid;HDCA:猪脱氧胆酸hyodeoxycholic acid;ω-MCA:ω-鼠胆酸omega-muricholic acid;MDCA:鼠脱氧胆酸murideoxycholic acid。 图 1 胆盐水解酶介导胆汁酸代谢 Fig. 1 BSH mediates bile acids metabolism[1, 12] |
微生物通常表达多种BSH基因,且其催化反应存在底物偏好性。例如,植物乳杆菌WCFS1、单核细胞增生李斯特氏菌等均表达多种BSH基因[17]。BSH可识别G-CBA和T-CBA,但是绝大多数微生物BSH对G-CBA存在偏好性,其对G-CBA水解效率强于T-CBA[18-21]。此外,部分肠道微生物BSH表现特定的胆汁酸去结合能力。例如,布氏乳杆菌JCM1069 BSH仅表现为牛磺脱氧胆酸水解酶活性,不具备牛磺胆酸水解酶活性[22];嗜酸乳杆菌NCFM BSH仅能水解含结合型鹅脱氧胆酸(chenodeoxycholic acid,CDCA)[23];植物乳杆菌BSH倾向于水解甘氨结合型脱氧胆酸,而动物乳杆菌BSH倾向于水解牛磺结合型脱氧胆酸[24]。
1.4 BSH分布最新研究指出,BSH基因广泛表达于12个门112个微生物属,包括乳杆菌属、双歧杆菌属、肠球菌属、梭菌属和拟杆菌属等肠道共生菌[25],其中59.73% BSH基因表达微生物属于厚壁菌门[26]。在肠道共生菌中,BSH基因还表达于单核细胞增生李斯特氏菌和牛布鲁氏菌等病原微生物[27-28]。此外,除了拟杆菌属和黄单胞菌属外,BSH基因表达菌均为革兰氏阳性菌,而包括埃希氏杆菌属和鼠伤寒沙门氏菌在内的革兰氏阴性菌既不具有BSH活性,也不表达BSH同源基因[29]。由于乳酸菌属具有人类和啮齿动物中最高的BSH活性,且其对7种主要CBA具有最稳定水解能力和最高活性[17, 26]。因此,当前对肠道微生物BSH研究集中于乳杆菌属。
2 BSH生物学功能BSH生物学功能主要从肠道微生物和宿主代谢2部分展开,见图 2。
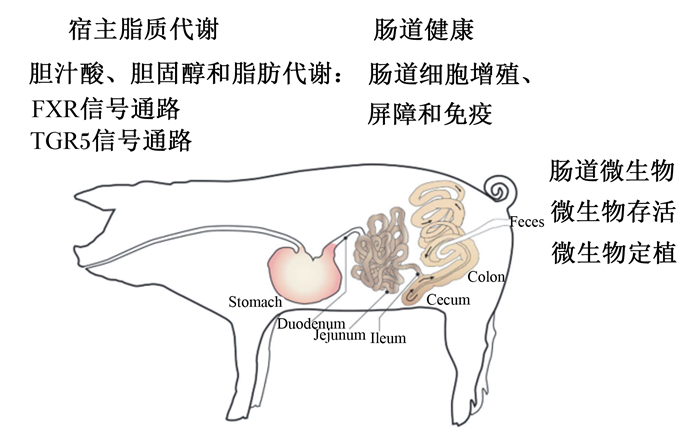
|
FXR:法尼醇X受体Farnesoid X receptor;TGR5:G蛋白偶联胆汁酸受体5 G protein coupled bile acid receptor 5;Stomach:胃;Duodenum:十二指肠;Jejunum:空肠;Ileum:回肠;Cecum:盲肠;Colon:结肠;Feces:粪便。 图 2 胆盐水解酶生物学功能(修改自参考文献[30]) Fig. 2 Biological functions of BSH (modified from reference [30]) |
包括乳酸菌在内众多微生物功能发挥有赖于其以活性形式定植于肠道,而BSH对微生物在肠道存活和定植均发挥重要作用。例如,与野生型相比,BSH基因突变单核细胞增生李斯特氏菌存活率显著降低[31];胃肠道中BSH基因重组植物乳杆菌和英诺克李斯特氏菌存活率均要显著高于野生型[32]。此外,BSH还与微生物在肠上皮细胞黏附作用密切相关。与BSH基因敲除植物乳杆菌相比,BSH基因表达植物乳杆菌对肠上皮细胞表现出更强的黏附能力[32]。目前关于BSH调控微生物存活和定植机制尚不清楚,已有研究主要围绕以下4方面进行[8, 33]:1)BSH的营养性作用。BSH介导去结合反应释放CBA中甘氨酸或牛磺酸,进而促进微生物生长。2)BSH改变微生物膜特性。微生物膜的组成、流动性、渗透性、疏水性和净电荷决定了微生物被宿主防御系统的损伤程度[34],BSH有助于胆固醇或胆汁酸并入细菌质膜,进而可能提高质膜的抗张强度以及改变其流动性和净电荷。此外,鉴于BSH酶也在致病微生物中表达以及BSH基因具有与水平基因转移作用,这可能对于病原微生物持续感染具有重要作用。3)BSH调节肠道微生物胆汁耐受。与野生型相比,BSH基因突变型植物乳杆菌和单核细胞增生李斯特氏菌均对胆盐更加敏感[24, 30];BSH调节微生物胆汁耐受功能与以下2方面有关,一方面,尽管UCBA被认为具有更强的细胞毒性,但其较低水溶性有助于降低其细胞毒性[17, 35],另一方面,UCBA通过结合微生物体内质子,缓解细胞内pH降低。4)肠道微生物BSH基因同系物。植物乳杆菌WCFS1和单核细胞增生李斯特氏菌等微生物中存在多种BSH基因同系物,BSH对CBA存在水解偏好可能有助于增强对其适应能力。
2.2 BSH调节宿主代谢 2.2.1 BSH参与调控宿主脂质代谢研究揭示,肠道微生物BSH具有调节宿主脂质代谢的功能[36]。直接证据表明,与野生型大肠杆菌组相比,BSH基因表达大肠杆菌显著降低小鼠体增重、脂肪沉积以及血清低密度脂蛋白和肝脏甘油三酯含量,血清低密度脂蛋白和肝脏甘油三酯含量降低幅度分别为60.6%和36.5%[3]。与此相似,BSH基因表达乳酸菌也对宿主脂质代谢具有调控作用。例如,乳酸杆菌可以显著降低肥胖人类和啮齿动物体脂含量、体脂百分比[37-38]以及血液低密度脂蛋白胆固醇、总胆固醇和非高密度脂蛋白胆固醇含量[39]。目前关于肠道微生物BSH与宿主脂质代谢直接关系的研究相对较少,有限证据表明其可能与以下2方面有关。
2.2.1.1 BSH通过肠道肠道法尼醇X受体(Farnesoid X receptor,FXR)信号调控宿主脂质代谢肠道FXR是一种核受体,广泛表达于肝脏和肠道等组织,对于宿主胆汁酸代谢、脂肪代谢、肠道屏障和炎症反应起到重要调节作用。例如,肠道FXR特异性抑制剂或茶褐素均通过抑制肠道FXR-Fgf15信号降低小鼠体增重、肝脏胆固醇含量和脂肪合成,其功能发挥主要通过抑制肠道微生物BSH活性,提高CBA水平[40-41]。与此相似,益生菌组合VSL#3提高肠道微生物BSH和胆汁酸去结合能力,进而抑制肠道FXR信号[42]。例外的是,与BSH基因敲除多形拟杆菌相比,野生型多形拟杆菌定植并未改变无菌小鼠肠道FXR信号通路[16],提示BSH功能与其微生物来源和种类有关。
2.2.1.2 BSH通过G蛋白偶联胆汁酸受体5(G protein coupled bile acid receptor 5,TGR5)信号调节宿主糖、脂代谢TGR5是G蛋白偶联受体家族的成员,广泛表达于胆管细胞、肠细胞、棕色脂肪组织、免疫细胞和肠道内分泌细胞等,在机体能量代谢中发挥重要调节作用[36]。一方面,TGR5通过激活肠道L细胞分泌胰高血糖素样肽-1(glucagon-like peptide-1,GLP-1),促进脂肪组织棕色化和提高胰岛素敏感性,缓解肥胖发生;另一方面,TGR5通过促进环腺苷酸释放,促进棕色脂肪组织产热。在天然胆汁酸中,次级胆汁酸LCA和DCA具有最高激活效力[36]。研究发现,植物乳杆菌通过降低回肠LCA和TGR5靶蛋白GLP-1,提高断奶仔猪血糖水平[43]。这提示BSH通过TGR5信号通路调控宿主糖、脂代谢。
2.2.2 BSH参与调控宿主肠道健康机体胆汁酸组成和水平受到肠道微生物调控,微生物BSH代谢产物次级胆汁酸对于肠道健康通常是有害的。例如,次级胆汁酸LCA和DCA通过诱导DNA氧化损伤和炎症等多种方式损伤肠道上皮细胞功能[4]。CBA对小肠上皮细胞增殖无作用或促进作用,UCBA则起到抑制小肠上皮细胞增殖作用[44]。在猪上进行的研究也得到一致结果,例如,次级胆汁酸(DCA和LCA)处理抑制猪小肠上皮细胞(IPEC-J2细胞)增殖和紧密连接蛋白表达[45]。次级胆汁酸HDCA抑制IPEC-J2细胞增殖[46],初级胆汁酸CDCA可以有效缓解脂多糖对IPEC-J2细胞肠道屏障的损伤[47]。
除肠道上皮细胞增殖之外,BSH代谢产物在调控肠道炎症中也发挥重要作用。肠道FXR信号缺失引起肠道炎症因子表达和淋巴结数量升高;而激活FXR通过稳定白细胞介素-1β(IL-1β)基因表达进而缓解肠道炎症[36]。相比于CBA,UCBA对FXR具有更强激活作用。Lin等[45]研究发现,次级胆汁酸LCA提高IPEC-J2细胞促炎因子白细胞介素-6(IL-6)和白细胞介素-8(IL-8)基因表达。因此,肠道微生物BSH可能通过介导初级胆汁酸向次级胆汁酸转化,进而损害肠道健康。
最近系列研究揭示,BSH代谢产物对宿主免疫具有调节作用。例如,DCA代谢物3β-羟基脱氧胆酸(3β-hydroxydeoxycholic acid,isoDCA)促进调节性T细胞产生[48];LCA代谢产物(3-oxoLCA和isoalloLCA)调节辅助性T细胞17和调节性T细胞的平衡进而调控宿主免疫反应机制[49],此外,isoalloLCA还可以高效抑制产毒型艰难梭菌在体内生长[50];粪菌移植可通过恢复肠道BSH活性治疗艰难梭状芽孢杆菌感染[51]。因此,肠道微生物BSH活性可作为人类疾病的无创监测指标[52]。本研究室研究发现,母猪和胎猪体内ω-MCA水平(2~148 nmol/L)远低于其发挥功能的水平,而并未检测出isoalloLCA[53],暗示其并未在母猪和胎猪上发挥免疫调节作用。
3 BSH抑制剂在畜牧业中潜在应用饲料中抗生素生长促进剂(antibiotic growth promoter,AGP)禁用是畜牧行业面临的巨大挑战之一。在当前“禁抗”背景下,如何开发和评价AGP替代产品迫在眉睫。尽管关于AGP调控动物生长作用机制还存在争论,但其对肠道微生物调控这一观点得到广泛认可[54]。胆汁酸代谢在AGP促生长发挥重要作用。研究揭示,AGP促生长作用与胆汁酸代谢、脂质代谢和肠道健康密切相关[55-57]。本研究室研究发现,脂多糖应激抑制断奶仔猪肠道FXR信号通路,三丁酸甘油酯通过恢复肠道FXR信号缓解脂多糖应激[58];最新研究指出,抗生素与氧化锌处理通过改变断奶仔猪胆汁酸代谢,激活肠道FXR和TGR5信号通路,维持肠道健康[56]。如前文所述,BSH具有抑制肠道FXR和TGR5功能,结合BSH抑制剂促进动物生长[59],这为BSH抑制剂在畜牧业中应用提供理论依据。
3.1 肠道微生物BSH与AGP早在1987年,Feighner等[60]发现BSH活性与禽类生长抑制密切相关。例如,饲粮中依罗霉素、维吉尼亚霉素等AGP与鸡生长性能和BSH活性之间密切关系[60],并指出BSH活性与禽生长抑制密切相关[61]。研究表明,AGP在提高动物生产性能的同时,降低胆汁酸去结合能力和BSH微生物表达丰度[62-64]。例如,Guban等[62]研究发现,杆菌肽处理提高肉鸡增重和饲料转化率,降低胆汁酸去结合能力,主要表现为CBA水平升高和BSH微生物丰度下降。Lin等[64]研究发现,饲粮添加泰乐菌素未改变肠道总菌量,却显著降低乳杆菌属表达丰度。
体外试验同样证实,抗生素对肠道微生物BSH活性具有抑制作用。例如,四环素类抗生素(土霉素、盐酸去甲金霉素、盐酸甲烯土霉素和盐酸强力霉素)、β-内酰胺类抗生素(邻氯青霉素和先锋霉素)、砷剂、林可酰胺类和磺胺类抗生素均可高效抑制BSH活性[65]。
3.2 BSH抑制剂研究进展Joyce等[3]采用表达BSH基因重组大肠杆菌,首次发现BSH对宿主体重的调控作用。在未改变采食量前提下,唾液乳酸杆菌JCM1046 BSH基因表达大肠杆菌(ECBSH1)体增重显著低于野生型大肠杆菌组,与对照组相比,ECBSH1组体重降低高达46%[3]。与上述相吻合,饮食中添加乳酸杆菌引起人类、啮齿动物、猪和禽类体重降低[37-38, 66-68]。Geng等[59]研究发现,饲粮添加BSH抑制剂显著提高肉鸡日增重。需要指出的是,并非所有肠道微生物BSH均发挥作用。例如,唾液乳酸杆菌UCC118 BSH并未发挥调控动物体重作用,罗伊氏乳杆菌I5007 BSH甚至有助于提高动物生长性能[3, 69]。导致差异原因可能与BSH底物特异性有关。例如,最新研究揭示,22种植物乳杆菌对猪胆汁酸去结合能力存在种内差异[35]。
基于BSH与动物生长和代谢之间密切联系,BSH抑制剂的开发逐渐引起动物营养学界的重视。例如,硫酸铜和硫酸锌均可抑制BSH活性,抑制率分别为91.7%和89.4%;硫酸锰、硫酸亚铁、碘酸钾和碘酸钠对BSH也具有强的抑制能力[70-72]。在此基础上,Smith等[65]构建BSH抑制剂高通量筛选系统,鉴定核黄素和咖啡酸苯乙酯等均为BSH高效抑制剂。作为BSH抑制剂,核黄素处理可以激活大鼠小肠上段和回肠FXR信号通路[73]。值得注意的是,肠道微生物BSH活性受到抑制在高脂饲粮引发的大鼠葡萄糖代谢紊乱和胰岛素抵抗中发挥重要作用[73],提示BSH抑制剂可能有助于提高机体血糖水平。此外,饲粮添加B族维生素以剂量依赖方式提高仔猪平均日增重和饲料转化效率[74]。最新研究发现,BSH抑制剂混合物(核黄素、咖啡酸苯乙酯和鼠尾草酸)显著提高科宝鸡平均日增重[57]。以上研究暗示BSH抑制剂在畜禽生产中具有广阔应用前景。
4 小结作为肠道微生物与宿主代谢之间纽带,肠道微生物代谢产物BSH通过介导肝脏来源的初级胆汁酸向非结合型胆汁酸以及次级胆汁酸转化,进而对微生物在肠道中存活和定植,宿主胆汁酸、胆固醇和脂肪代谢以及肠道上皮细胞增殖、屏障、炎症和免疫等多方面发挥调控作用。值得重视的是,对于人类患者,一方面,BSH通过FXR和TGR5信号通路促进脂肪分解,提高胰岛素敏感性和抑制脂肪合成,维持肥胖病人脂质代谢稳态;另一方面,BSH部分代谢产物次级胆汁酸(例如isoalloLCA)通过调节辅助性T细胞17和调节性T细胞的平衡,进而调节宿主的免疫功能。母猪和胎猪体内低表达isoalloLCA,暗示其可能并未有效调节宿主的免疫功能。对于畜禽动物,高BSH活性可能不利于动物最大生长潜力的发挥,通过筛选并应用肠道微生物BSH抑制剂,可能有助于实现其最佳生长速度、饲料转化效率和肠道健康。此外,目前在畜牧生产中广泛使用膳食纤维、益生菌和酸化剂等产品,其功能的发挥与肠道微生物密切相关,但是其与肠道微生物BSH之间联系尚不明确。因此,如何基于BSH开发和评价益生菌、益生元以及其他AGP替代产品是未来重要研究方向。
| [1] |
PERINO A, DEMAGNY H, VELAZQUEZ-VILLEGAS L, et al. Molecular physiology of bile acid signaling in health, disease, and aging[J]. Physiological Reviews, 2021, 101(2): 683-731. DOI:10.1152/physrev.00049.2019 |
| [2] |
FOLEY M H, O'FLAHERTY S, ALLEN G, et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(6): e2017709118. DOI:10.1073/pnas.2017709118 |
| [3] |
JOYCE S A, MACSHARRY J, CASEY P G, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(20): 7421-7426. DOI:10.1073/pnas.1323599111 |
| [4] |
JIA W, XIE G X, JIA W P. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis[J]. Nature Reviews.Gastroenterology & Hepatology, 2018, 15(2): 111-128. |
| [5] |
KIM G B, MIYAMOTO C M, MEIGHEN E A, et al. Cloning and characterization of the bile salt hydrolase genes (BSH) from Bifidobacterium bifidum strains[J]. Applied and Environmental Microbiology, 2004, 70(9): 5603-5612. DOI:10.1128/AEM.70.9.5603-5612.2004 |
| [6] |
GOPAL-SRIVASTAVA R, HYLEMON P B. Purification and characterization of bile salt hydrolase from Clostridium perfringens[J]. Journal of Lipid Research, 1988, 29(8): 1079-1085. DOI:10.1016/S0022-2275(20)38464-9 |
| [7] |
GRILL J, SCHNEIDER F, CROCIANI J, et al. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536[J]. Applied and Environmental Microbiology, 1995, 61(7): 2577-2582. DOI:10.1128/aem.61.7.2577-2582.1995 |
| [8] |
BEGLEY M, HILL C, GAHAN C G M. Bile salt hydrolase activity in probiotics[J]. Applied and Environmental Microbiology, 2006, 72(3): 1729-1738. DOI:10.1128/AEM.72.3.1729-1738.2006 |
| [9] |
CHIANG J Y L. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms[J]. Journal of Hepatology, 2004, 40(3): 539-551. DOI:10.1016/j.jhep.2003.11.006 |
| [10] |
RUSSELL D W. The enzymes, regulation, and genetics of bile acid synthesis[J]. Annual Review of Biochemistry, 2003, 72: 137-174. DOI:10.1146/annurev.biochem.72.121801.161712 |
| [11] |
GARCÍA-CAÑAVERAS J C, DONATO M T, CASTELL J V, et al. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method[J]. Journal of Lipid Research, 2012, 53(10): 2231-2241. DOI:10.1194/jlr.D028803 |
| [12] |
WANG P, ZHONG H J, SONG Y M, et al. Targeted metabolomics analysis of maternal-placental-fetal metabolism in pregnant swine reveals links in fetal bile acid homeostasis and sulfation capacity[J]. American Journal of Physiology: Gastrointestinal and Liver Physiology, 2019, 317(1): G8-G16. DOI:10.1152/ajpgi.00056.2019 |
| [13] |
RIDLON J M, KANG D J, HYLEMON P B. Bile salt biotransformations by human intestinal bacteria[J]. Journal of Lipid Research, 2006, 47(2): 241-259. |
| [14] |
PARASAR B, ZHOU H, XIAO X Y, et al. Chemoproteomic profiling of gut microbiota-associated bile salt hydrolase activity[J]. ACS Central Science, 2019, 5(5): 867-873. DOI:10.1021/acscentsci.9b00147 |
| [15] |
SAYIN S I, WAHLSTRÖM A, FELIN J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist[J]. Cell Metabolism, 2013, 17(2): 225-235. DOI:10.1016/j.cmet.2013.01.003 |
| [16] |
YAO L N, SEATON S C, NDOUSSE-FETTER S, et al. A selective gut bacterial bile salt hydrolase alters host metabolism[J]. eLife, 2018, 7: e37182. DOI:10.7554/eLife.37182 |
| [17] |
CHAND D, AVINASH V S, YADAV Y, et al. Molecular features of bile salt hydrolases and relevance in human health[J]. Biochimica et Biophysica Acta: General Subjects, 2017, 1861(1 Pt A): 2981-2991. |
| [18] |
KIM G B, YI S H, LEE B H. Purification and characterization of three different types of bile salt hydrolases from Bifidobacterium strains[J]. Journal of Dairy Science, 2004, 87(2): 258-266. DOI:10.3168/jds.S0022-0302(04)73164-1 |
| [19] |
COLEMAN J P, HUDSON L L. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens[J]. Applied and Environmental Microbiology, 1995, 61(7): 2514-2520. DOI:10.1128/aem.61.7.2514-2520.1995 |
| [20] |
TANAKA H, HASHIBA H, KOK J, et al. Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization[J]. Applied and Environmental Microbiology, 2000, 66(6): 2502-2512. DOI:10.1128/AEM.66.6.2502-2512.2000 |
| [21] |
TARANTO M P, SESMA F, DE VALDEZ G F. Localization and primary characterization of bile salt hydrolase from Lactobacillus reuteri[J]. Biotechnology Letters, 1999, 21(11): 935-938. DOI:10.1023/A:1005652501404 |
| [22] |
MOSER S A, SAVAGE D C. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in Lactobacilli[J]. Applied and Environmental Microbiology, 2001, 67(8): 3476-3480. DOI:10.1128/AEM.67.8.3476-3480.2001 |
| [23] |
MCAULIFFE O, CANO R J, KLAENHAMMER T R. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM[J]. Applied and Environmental Microbiology, 2005, 71(8): 4925-4929. DOI:10.1128/AEM.71.8.4925-4929.2005 |
| [24] |
DE SMET I, VAN HOORDE L, VANDE WOESTYNE M, et al. Significance of bile salt hydrolytic activities of Lactobacilli[J]. The Journal of Applied Bacteriology, 1995, 79(3): 292-301. DOI:10.1111/j.1365-2672.1995.tb03140.x |
| [25] |
JONES B V, BEGLEY M, HILL C, et al. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(36): 13580-13585. DOI:10.1073/pnas.0804437105 |
| [26] |
SONG Z W, CAI Y Y, LAO X Z, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome[J]. Microbiome, 2019, 7(1): 9. DOI:10.1186/s40168-019-0628-3 |
| [27] |
BUSTOS A Y, FONT DE VALDEZ G, FADDA S, et al. New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health[J]. Food Research International, 2018, 112: 250-262. DOI:10.1016/j.foodres.2018.06.035 |
| [28] |
MARCHESINI M I, CONNOLLY J, DELPINO M V, et al. Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization[J]. PLoS One, 2011, 6(12): e28480. DOI:10.1371/journal.pone.0028480 |
| [29] |
DEAN M, CERVELLATI C, CASANOVA E, et al. Characterization of cholylglycine hydrolase from a bile-adapted strain of Xanthomonas maltophilia and its application for quantitative hydrolysis of conjugated bile salts[J]. Applied and Environmental Microbiology, 2002, 68(6): 3126-3128. DOI:10.1128/AEM.68.6.3126-3128.2002 |
| [30] |
HOLMAN D B, BRUNELLE B W, TRACHSEL J, et al. Meta-analysis to define a core microbiota in the swine gut[J]. mSystems, 2017, 2(3): e00004-17. DOI:10.1128/mSystems.00004-17 |
| [31] |
BEGLEY M, SLEATOR R D, GAHAN C G M, et al. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes[J]. Infection and Immunity, 2005, 73(2): 894-904. DOI:10.1128/IAI.73.2.894-904.2005 |
| [32] |
YANG Y, LIU Y R, ZHOU S S, et al. Bile salt hydrolase can improve Lactobacillus plantarum survival in gastrointestinal tract by enhancing their adhesion ability[J]. FEMS Microbiology Letters, 2019, 366(8): fnz100. DOI:10.1093/femsle/fnz100 |
| [33] |
INAGAKI T, MOSCHETTA A, LEE Y K, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(10): 3920-3925. DOI:10.1073/pnas.0509592103 |
| [34] |
PESCHEL A. How do bacteria resist human antimicrobial peptides?[J]. Trends in Microbiology, 2002, 10(4): 179-186. DOI:10.1016/S0966-842X(02)02333-8 |
| [35] |
PRETE R, LONG S L, GALLARDO A L, et al. Beneficial bile acid metabolism from Lactobacillus plantarum of food origin[J]. Scientific Reports, 2020, 10(1): 1165. DOI:10.1038/s41598-020-58069-5 |
| [36] |
SUN L L, CAI J, GONZALEZ F J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer[J]. Nature Reviews.Gastroenterology & Hepatology, 2021, 18(5): 335-347. |
| [37] |
JUNG S, LEE Y J, KIM M, et al. Supplementation with two probiotic strains, Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032, reduced body adiposity and Lp-PLA2 activity in overweight subjects[J]. Journal of Functional Foods, 2015, 19(Part A): 744-752. |
| [38] |
KIM M, KIM M, KANG M, et al. Effects of weight loss using supplementation with Lactobacillus strains on body fat and medium-chain acylcarnitines in overweight individuals[J]. Food & Function, 2017, 8(1): 250-261. |
| [39] |
JONES M L, MARTONI C J, PARENT M, et al. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults[J]. The British Journal of Nutrition, 2012, 107(10): 1505-1513. DOI:10.1017/S0007114511004703 |
| [40] |
HUANG F J, ZHENG X J, MA X H, et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism[J]. Nature Communications, 2019, 10(1): 4971. DOI:10.1038/s41467-019-12896-x |
| [41] |
JIANG C T, XIE C, LV Y, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction[J]. Nature Communications, 2015, 6: 10166. DOI:10.1038/ncomms10166 |
| [42] |
DEGIROLAMO C, RAINALDI S, BOVENGA F, et al. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice[J]. Cell Reports, 2014, 7(1): 12-18. DOI:10.1016/j.celrep.2014.02.032 |
| [43] |
LIN S, YANG X M, LONG Y R, et al. Dietary supplementation with Lactobacillus plantarum modified gut microbiota, bile acid profile and glucose homoeostasis in weaning piglets[J]. The British Journal of Nutrition, 2020, 124(8): 797-808. DOI:10.1017/S0007114520001774 |
| [44] |
DOSSA A Y, ESCOBAR O, GOLDEN J, et al. Bile acids regulate intestinal cell proliferation by modulating EGFR and FXR signaling[J]. American Journal of Physiology.Gastrointestinal and Liver Physiology, 2016, 310(2): G81-G92. DOI:10.1152/ajpgi.00065.2015 |
| [45] |
LIN S, YANG X M, YUAN P Q, et al. Undernutrition shapes the gut microbiota and bile acid profile in association with altered gut-liver FXR signaling in weaning pigs[J]. Journal of Agricultural and Food Chemistry, 2019, 67(13): 3691-3701. DOI:10.1021/acs.jafc.9b01332 |
| [46] |
SONG M, YANG Q, ZHANG F L, et al. Hyodeoxycholic acid (HDCA) suppresses intestinal epithelial cell proliferation through FXR-PI3K/AKT pathway, accompanied by alteration of bile acids metabolism profiles induced by gut bacteria[J]. FASEB Journal, 2020, 34(5): 7103-7117. DOI:10.1096/fj.201903244R |
| [47] |
SONG M, YE J Y, ZHANG F L, et al. Chenodeoxycholic acid (CDCA) protects against the lipopolysaccharide-induced impairment of the intestinal epithelial barrier function via the FXR-MLCK pathway[J]. Journal of Agricultural and Food Chemistry, 2019, 67(32): 8868-8874. DOI:10.1021/acs.jafc.9b03173 |
| [48] |
CAMPBELL C, MCKENNEY P T, KONSTANTINOVSKY D, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells[J]. Nature, 2020, 581(7809): 475-479. DOI:10.1038/s41586-020-2193-0 |
| [49] |
HANG S Y, PAIK D, YAO L N, et al. Bile acid metabolites control TH17 and Treg cell differentiation[J]. Nature, 2019, 576(7785): 143-148. DOI:10.1038/s41586-019-1785-z |
| [50] |
SATO Y, ATARASHI K, PLICHTA D R, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians[J/OL]. Nature, 2021. (2021-07-29)[2021-08-23]. https://doi.org/10.1038/s41586-021-03832-5.DOI: 10.1038/s41586-021-03832-5
|
| [51] |
MULLISH B H, MCDONALD J A K, PECHLIVANIS A, et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection[J]. Gut, 2019, 68(10): 1791-1800. DOI:10.1136/gutjnl-2018-317842 |
| [52] |
KHODAKIVSKYI P V, LAUBER C L, YEVTODIYENKO A, et al. Noninvasive imaging and quantification of bile salt hydrolase activity: from bacteria to humans[J]. Science Advances, 2021, 7(6): eaaz9857. DOI:10.1126/sciadv.aaz9857 |
| [53] |
王朋. 母猪妊娠期胆汁酸代谢对胎猪存活的影响及其调控[D]. 博士学位论文. 成都: 四川农业大学, 2020. WANG P. Effects of bile acids metabolism in sows during pregnancy on fetal pigs survival and their regulation[D]. Ph. D. Thesis. Chengdu: Sichuan Agricultural University, 2020. (in Chinese) |
| [54] |
GASKINS H R, COLLIER C T, ANDERSON D B. Antibiotics as growth promotants: mode of action[J]. Animal Biotechnology, 2002, 13(1): 29-42. DOI:10.1081/ABIO-120005768 |
| [55] |
HU Y, ZHANG Y H, LIU C, et al. Multi-omics profiling highlights lipid metabolism alterations in pigs fed low-dose antibiotics[J]. BMC Genetics, 2020, 21(1): 112. DOI:10.1186/s12863-020-00918-3 |
| [56] |
IPHARRAGUERRE I R, PASTOR J J, GAVALDÀ-NAVARRO A, et al. Antimicrobial promotion of pig growth is associated with tissue-specific remodeling of bile acid signature and signaling[J]. Scientific Reports, 2018, 8(1): 13671. DOI:10.1038/s41598-018-32107-9 |
| [57] |
CHO I, YAMANISHI S, COX L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity[J]. Nature, 2012, 488(7413): 621-626. DOI:10.1038/nature11400 |
| [58] |
GU Y, SONG Y, YIN H, et al. Dietary supplementation with tributyrin prevented weaned pigs from growth retardation and lethal infection via modulation of inflammatory cytokines production, ileal FGF19 expression, and intestinal acetate fermentation[J]. Journal of Animal Science, 2017, 95(1): 226-238. |
| [59] |
GENG W J, LONG S L, CHANG Y J, et al. Evaluation of bile salt hydrolase inhibitor efficacy for modulating host bile profile and physiology using a chicken model system[J]. Scientific Reports, 2020, 10(1): 4941. DOI:10.1038/s41598-020-61723-7 |
| [60] |
FEIGHNER S D, DASHKEVICZ M P. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity[J]. Applied and Environmental Microbiology, 1987, 53(2): 331-336. DOI:10.1128/aem.53.2.331-336.1987 |
| [61] |
FEIGHNER S D, DASHKEVICZ M P. Effect of dietary carbohydrates on bacterial cholyltaurine hydrolase in poultry intestinal homogenates[J]. Applied and Environmental Microbiology, 1988, 54(2): 337-342. DOI:10.1128/aem.54.2.337-342.1988 |
| [62] |
GUBAN J, KORVER D R, ALLISON G E, et al. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens[J]. Poultry Science, 2006, 85(12): 2186-2194. DOI:10.1093/ps/85.12.2186 |
| [63] |
KNARREBORG A, LAURIDSEN C, ENGBERG R M, et al. Dietary antibiotic growth promoters enhance the bioavailability of alpha-tocopheryl acetate in broilers by altering lipid absorption[J]. The Journal of Nutrition, 2004, 134(6): 1487-1492. DOI:10.1093/jn/134.6.1487 |
| [64] |
LIN J, HUNKAPILLER A A, LAYTON A C, et al. Response of intestinal microbiota to antibiotic growth promoters in chickens[J]. Foodborne Pathogens and Disease, 2013, 10(4): 331-337. DOI:10.1089/fpd.2012.1348 |
| [65] |
SMITH K, ZENG X M, LIN J. Discovery of bile salt hydrolase inhibitors using an efficient high-throughput screening system[J]. PLoS One, 2014, 9(1): e85344. DOI:10.1371/journal.pone.0085344 |
| [66] |
HEENEY D D, ZHAI Z Y, BENDIKS Z, et al. Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro[J]. Gut Microbes, 2019, 10(3): 382-397. DOI:10.1080/19490976.2018.1534513 |
| [67] |
SHARIFI S D, DIBAMEHR A, LOTFOLLAHIAN H, et al. Effects of flavomycin and probiotic supplementation to diets containing different sources of fat on growth performance, intestinal morphology, apparent metabolizable energy, and fat digestibility in broiler chickens[J]. Poultry Science, 2012, 91(4): 918-927. DOI:10.3382/ps.2011-01844 |
| [68] |
RIBOULET-BISSON E, STURME M H J, JEFFERY I B, et al. Effect of Lactobacillus salivarius bacteriocin Abp118 on the mouse and pig intestinal microbiota[J]. PLoS One, 2012, 7(2): e31113. DOI:10.1371/journal.pone.0031113 |
| [69] |
HOU C L, ZENG X F, YANG F J, et al. Study and use of the probiotic Lactobacillus reuteri in pigs: a review[J]. Journal of Animal Science and Biotechnology, 2015, 6(1): 14. DOI:10.1186/s40104-015-0014-3 |
| [70] |
WANG Z, ZENG X M, MO Y M, et al. Identification and characterization of a bile salt hydrolase from Lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters[J]. Applied and Environmental Microbiology, 2012, 78(24): 8795-8802. DOI:10.1128/AEM.02519-12 |
| [71] |
JACELA J Y, DEROUCHEY J M, TOKACH M D, et al. Feed additives for swine: fact sheets-high dietary levels of copper and zinc for young pigs, and phytase[J]. Journal of Swine Health and Production, 2010, 18(2): 87-91. |
| [72] |
SHELTON N W, TOKACH M D, NELSSEN J L, et al. Effects of copper sulfate, tri-basic copper chloride, and zinc oxide on weanling pig performance[J]. Journal of Animal Science, 2011, 89(8): 2440-2451. DOI:10.2527/jas.2010-3432 |
| [73] |
WAISE T M Z, LIM Y M, DANAEI Z, et al. Small intestinal taurochenodeoxycholic acid-FXR axis alters local nutrient-sensing glucoregulatory pathways in rats[J]. Molecular Metabolism, 2021, 44: 101132. DOI:10.1016/j.molmet.2020.101132 |
| [74] |
STAHLY T S, WILLIAMS N H, LUTZ T R, et al. Dietary B vitamin needs of strains of pigs with high and moderate lean growth[J]. Journal of Animal Science, 2007, 85(1): 188-195. DOI:10.2527/jas.2006-086 |




