2. 塔里木畜牧科技兵团重点实验室, 阿拉尔 843300;
3. 南京农业大学动物科技学院, 南京 210032
2. Key Laboratory of Tarim Animal Science and Technology Corps, Alar 843300, China;
3. College of Animal Science and Technology, Nanjing Agricultural University, Nanjing 210032, China
新疆是我国的畜牧大省,有着丰富的物种资源,拥有超过3 000万只的本地品种羊资源,主要有策勒黑羊、和田羊、卡拉库尔羊、哈萨克羊、多浪羊等品种,这些本地品种羊有着耐粗饲和对环境适应性强等优点[1-2]。近几年新疆响应国家号召大力发展畜牧业,从国内外引进了大量的优秀品种来扩大繁育,湖羊因其生长发育快、多胎性等特点成为了新疆引种扩繁的重点[3-4]。瘤胃是反刍动物重要的消化器官,内部有丰富的瘤胃微生物,瘤胃菌群的结构处于相对稳定的动态平衡,该平衡对于维持动物机体健康、瘤胃内环境稳定、营养物质消化和吸收及提高动物的生产性能等起着重要的作用[5-7]。瘤胃菌群结构主要受动物品种、日龄、地区、饲粮结构等多种因素的影响[8-10]。不同品种之间的瘤胃菌群结构具有一定的相似性,但其菌群组成和相对丰度存在一定差异。King等[11]研究发现,在相同的饲养条件下,娟姗牛和荷斯坦牛瘤胃菌群结构相似度很高,但娟姗牛瘤胃内产甲烷短杆菌的相对丰度显著高于荷斯坦牛。韩帅[12]在比较呼伦贝尔羔羊和呼伦贝尔羊与杜泊羊杂一代(呼杜杂一代)羔羊瘤胃微生物动态的研究中发现呼杜杂一代羔羊在产甲烷菌、脂解厌氧弧杆菌、反刍月形单胞菌、牛链球菌的相对丰度上高于呼伦贝尔羔羊。以上研究表明不同品种间由于动物品种遗传学差异会引起瘤胃内部分菌落相对丰度的差异。本试验以新疆地方品种羊卡拉库尔羊和引入品种湖羊为研究对象,在饲喂相同饲粮的条件下分析湖羊与卡拉库尔羊瘤胃菌群结构的差异性,揭示卡拉库尔羊较湖羊耐受高粗饲料饲粮的瘤胃微生物机制,为南疆地区不同品种羊的饲养管理提供参考依据。
1 材料与方法 1.1 试验设计与饲养管理本试验选取体重[(35±2) kg]相近的健康卡拉库尔羊羯羊和湖羊羯羊各6只,饲喂相同饲粮。参考《肉羊饲养标准》(NY/T 816—2004)[13]配制基础饲粮(维持需要),基础饲粮组成及营养水平如表 1所示,分别于每日08:00与20:00分2次限量饲喂1.2 kg,自由饮水。
|
|
表 1 基础饲粮组成及营养水平(风干基础) Table 1 Composition and nutrient levels of the basal diet (air-dry basis) |
试验预试期为30 d,正试期为3 d。在正试期第1~3天于晨饲前通过口腔采集瘤胃液,经4层纱布过滤,每天收集每组混合瘤胃液约50 mL,立即放入液氮中,后于-80 ℃冰箱保存,以备送检。
1.3 样品送检将瘤胃液样品送至百迈客生物有限公司用PacBio测序方法对细菌16S rRNA基因进行高通量测序。
1.4 测序及生物信息分析提取样品总DNA后,利用细菌16S rRNA基因全长引物序列合成带有Barcode的特异引物,进行PCR扩增并对其产物进行纯化、定量和均一化形成测序文库(SMRT Bell),建好的文库先进行文库质检,质检合格的文库用PacBio Sequel进行测序。通过smrtlink分析软件导出CCS文件,根据Barcode序列识别不同样品的数据并转化为fastq格式数据。数据预处理:将PacBio下机的数据导出为CCS文件(CCS序列使用Pacbio提供的smrtlink工具获取)后,主要有如下3个步骤:1)CCS识别。使用lima v1.7.0软件,通过Barcode对CCS进行识别,得到的Raw-CCS序列数据。2)CCS过滤。使用cutadapt 1.9.1软件进行引物序列的识别与去除并且进行长度过滤,得到不包含引物序列的Clean-CCS序列。3)去除嵌合体。使用UCHIME v4.2软件,鉴定并去除嵌合体序列,得到Effective-CCS序列。后续进行多样性分析、差异分析、相关性分析及功能预测等。
1.5 数据处理试验数据用SPSS 26.0统计分析软件进行独立样本t检验,P>0.05表示差异不显著,P<0.05表示差异显著,P<0.01表示差异极显著,试验结果用“平均值±标准差”表示。
2 结果与分析 2.1 湖羊与卡拉库尔羊瘤胃菌群Alpha多样性分析由表 2可知,样品的覆盖度(Coverage)均约等于1,说明本次测序结果能够反映样本的真实情况;卡拉库尔羊的操作分类单元(OTUs)数目、ACE指数、Chao1指数、Shannon指数、Simpson指数均极显著高于湖羊(P<0.01),说明在饲喂相同饲粮条件下卡拉库尔羊瘤胃菌群的多样性和丰富度均高于湖羊。
|
|
表 2 湖羊与卡拉库尔羊瘤胃菌群Alpha多样性指数 Table 2 Alpha diversity indices of rumen microflora in Hu sheep and Karakul sheep |
如图 1所示,本试验所有样本共得到626个OTUs,其中278个OTUs为2个品种羊共有的,占OTUs总数的44.4%。湖羊特有OTUs 109个,占17.4%;卡拉库尔羊特有OTUs 239个,占38.2%,上述结果说明湖羊和卡拉库尔羊的瘤胃菌群结构存在差异性。
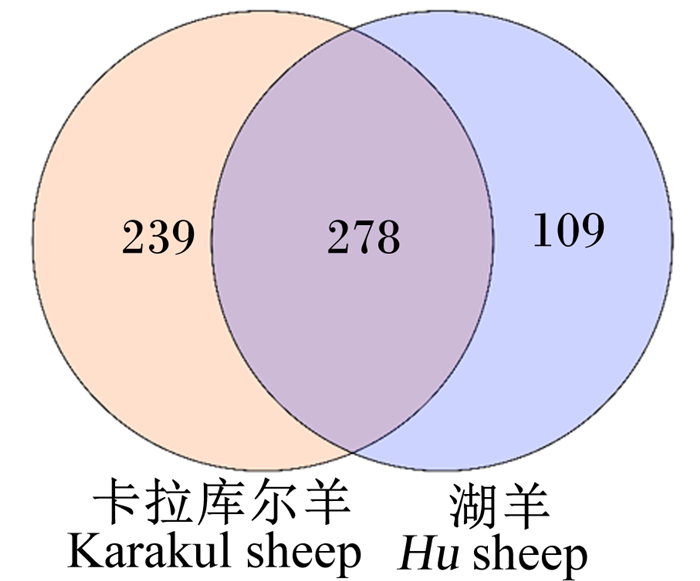
|
图 1 维恩图 Fig. 1 Venn graph |
湖羊和卡拉库尔羊瘤胃菌群在门水平上的组成和相对丰度如表 3所示。本试验共获得20个细菌门,其中在至少1个品种羊上的相对丰度大于1%的菌门有厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)、变形菌门(Proteobacteria)、蓝细菌门(Cyanobacteria)、Kiritimatiellaeota、软壁菌门(Tenericutes)、浮霉菌门(Planctomycetes)、髌骨细菌门(Patescibacteria)和互养菌门(Synergistetes)。湖羊的优势菌门为厚壁菌门(86.25%)、拟杆菌门(4.3%)和蓝细菌门(3.12%);卡拉库尔羊的优势菌门为厚壁菌门(59.57%)、拟杆菌门(21.34%)和Kiritimatiellaeota(9.75%)。湖羊厚壁菌门和蓝细菌门的相对丰度显著高于卡拉库尔羊(P < 0.05),髌骨细菌门和互养菌门的相对丰度显著低于卡拉库尔羊(P < 0.05),拟杆菌门、变形菌门、Kiritimatiellaeota和软壁菌门的相对丰度极显著低于卡拉库尔羊(P < 0.01)。
|
|
表 3 湖羊与卡拉库尔羊瘤胃菌群在门水平上的组成和相对丰度 Table 3 Composition and relative abundances of rumen microflora at phylum level in Hu sheep and Karakul sheep |
湖羊和卡拉库尔羊瘤胃菌群在属水平上的组成和相对丰度如表 4所示。本试验在属水平上共鉴定出130个细菌属,在至少1个品种羊中相对丰度大于1%的菌属有10个。湖羊的优势菌属有为奎恩氏菌属(46.43%)、丁酸弧菌属(6.34%)和克里斯滕森菌科_R-7_群(5.72%);卡拉库尔羊的优势菌属为普雷沃氏菌属_1(18.20%)、克里斯滕森菌科_R-7_群(17.09%)和奎恩氏菌属(6.97%)。湖羊奎恩氏菌属的相对丰度极显著高于卡拉库尔羊(P < 0.01),丁酸弧菌属的相对丰度显著高于卡拉库尔羊(P < 0.05);卡拉库尔羊克里斯滕森菌科_R-7_群、普雷沃氏菌属_1、瘤胃球菌科_NK4A214、Uncultured_bacterium_o_WCHB1-41、Uncultured_bacterium_o_Mollicutes_RF39的相对丰度极显著高于湖羊(P < 0.01),毛螺旋菌科_XPB1014_群、解琥珀酸菌属、瘤胃球菌科_UCG-014的相对丰度显著高于湖羊(P < 0.05)。
|
|
表 4 湖羊与卡拉库尔羊瘤胃细菌菌群在属水平上的相对丰度 Table 4 Composition and relative abundances of rumen microflora at genus level in Hu sheep and Karakul sheep |
湖羊在细胞过程和环境信息处理2个生物代谢通路的富集高于卡拉库尔羊(图 2);卡拉库尔羊在生物系统和遗传信息处理2个生物代谢通路的富集高于湖羊。进一步对预测基因二级功能层进行分析(图 3),发现湖羊在传染病(细菌)、细胞运动、辅因子和维生素的代谢、能量代谢以及折叠、修饰和降解等生物代谢通路的富集高于卡拉库尔羊;而卡拉库尔羊在核苷酸代谢、脂质代谢、复制和修复、氨基酸代谢、其他次生代谢物的生物合成、碳水化合物代谢、翻译、老化、膜运输等方面生物代谢通路的富集高于湖羊。
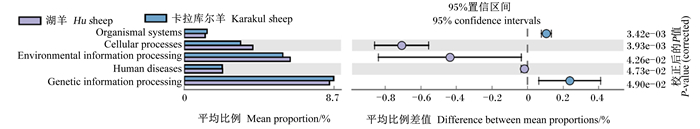
|
Organismal systems:生物系统;Cellular processes:细胞过程;Environmental information processing:环境信息处理;Human diseases:人类疾病;Genetic information processing:遗传信息处理。 图 2 湖羊与卡拉库尔羊生物代谢通路功能(KEGG通路1级) Fig. 2 Biological metabolism pathway function in Hu sheep and Karakul sheep (KEGG pathway class 1) |
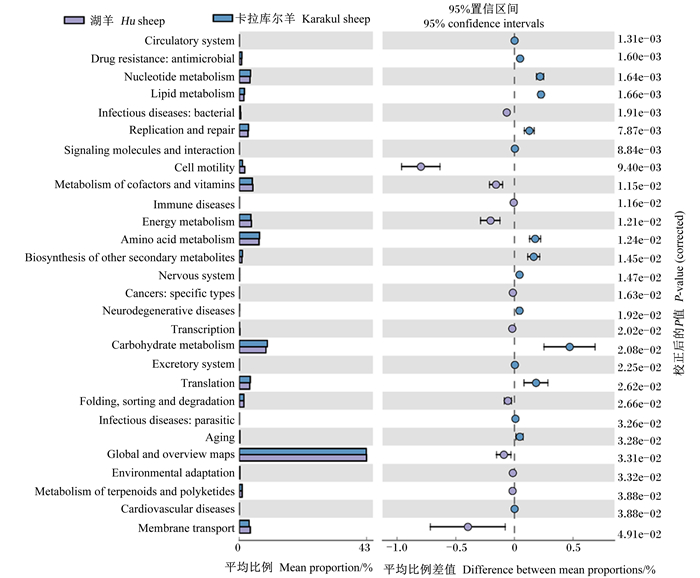
|
Circulatory system:循环系统;Drug resistance: antimicrobial:耐药性(抗菌);Nucleotide metabolism:核苷酸代谢;Lipid metabolism:脂质代谢;Infectious diseases: bacterial:传染病(细菌);Replication and repair:复制和修复;Signaling molecules and interaction:信号分子和相互作用;细胞运动:Cell motility;辅因子和维生素的代谢:Metabolism of cofactors and vitamins;Immune diseases:免疫疾病;Energy metabolism:能量代谢;Amino acid metabolism:氨基酸代谢;Biosynthesis of other secondary metabolites:其他次生代谢物的生物合成;Nervous system:神经系统;Cancers: specific types:癌症:特定类型;Neurodegenerative diseases:神经退行性疾病;Transcription:转录;Carbohydrate metabolism:碳水化合物代谢;Excretory system:排泄系统;Translation:翻译;Folding, sorting and degradation:折叠、分类和降解;Infectious diseases: parasitic:传染病(寄生虫);Aging:老化;Global and overview maps:全局和概览图;Environmental adaptation:环境适应;Metabolism of terpenoids and polyketides:萜类化合物和聚酮化合物的代谢;Cardiovascular diseases:心血管疾病;Membrane transport:膜运输。 图 3 湖羊与卡拉库尔羊生物代谢通路功能(KEGG通路2级) Fig. 3 Biological metabolism pathway function in Hu sheep and Karakul sheep (KEGG pathway class 2) |
瘤胃微生物对反刍动物的生产性能、营养物质消化吸收有着重要的意义。动物品种被证明是影响反刍动物瘤胃菌群结构的重要因素[14]。Chang等[15]研究表明,藏绵羊的瘤胃菌群丰富度和多样性均显著高于小尾寒羊和杜波羊。Huang等[16]报道,西藏绵羊比甘肃高山细毛羊更能适应高原恶劣环境的原因可能是西藏绵羊瘤胃菌群组成更多样性。本研究中卡拉库尔羊的瘤胃菌群丰富度和多样性均高于湖羊,暗示卡拉库尔羊对环境拥有更好的适应能力。
在本研究中湖羊与卡拉库尔羊的优势菌门均为厚壁菌门和拟杆菌门,且厚壁菌门的相对丰度高于拟杆菌门。这与众多研究描述的瘤胃内厚壁菌门的相对丰度高于拟杆菌门的结果[17-18]相一致,但与一些研究描述的拟杆菌门相对丰度高于厚壁菌门的报道[19-20]不同。这可能与羊的品种、饲粮结构及饲养环境的差异有关。厚壁菌门和拟杆菌门被报道是瘤胃内最丰富的菌门[21],其中厚壁菌门主要与瘤胃内纤维的降解和脂肪的沉积有关[22-23],而拟杆菌门主要参与淀粉的降解过程[21]。Fernando等[24]研究发现,牛瘤胃菌群中厚壁菌门与拟杆菌门的比值随着饲粮结构中粗饲料占比的增加而增加。本研究中湖羊与卡拉库尔羊优势菌门中厚壁菌门的相对丰度更高,这与本试验采用的是高粗饲料占比的饲粮结构相一致。
在本试验中湖羊的优势菌属是奎恩氏菌属、丁酸弧菌属和克里斯滕森菌属_R-7_群;卡拉库尔羊的优势菌属为普雷沃氏菌属_1、克里斯滕森菌属_R-7_群和奎恩氏菌属。奎恩氏菌属被报道主要参与瘤胃内碳水化合物的降解过程[25],本试验中湖羊奎恩氏菌属的相对丰度极显著高于卡拉库尔羊,表明湖羊可能在消化富含非纤维碳水化合物的饲粮时更具优势。克里斯滕森菌科_R-7_群是参与蛋白质降解过程的主要菌属[26]。普雷沃氏菌和丁酸弧菌被证明与瘤胃内纤维的降解过程相关[27-29]。其中普雷沃氏菌属于拟杆菌门,是瘤胃内最活跃的菌属,不仅参与纤维素的降解过程,还与瘤胃内蛋白质、可溶性碳水化合物和半纤维素的降解相关[30-31]。提高饲粮中精料的占比会引起普雷沃氏菌相对丰度的增加[32],而当饲粮结构中粗饲料占比高时也会引起普雷沃氏菌的大量增殖[33]。其原因可能是普雷沃氏菌既参与蛋白质和碳水化合物的降解和过程,又是瘤胃内主要的纤维素菌。卡拉库尔羊的普雷沃氏菌属_1的相对丰度极显著高于湖羊,这或许是卡拉库尔羊具有更强适应性的原因。本试验未能开展更多品种和饲粮结构对反刍动物瘤胃菌群影响的综合研究,具有一定的局限性,因此,后期需要综合研究品种和饲粮结构对反刍动物瘤胃菌群的影响作用。
KEGG代谢途径常用来分析微生物群落的功能基因在代谢途径上的差异,是研究群落样本为适应环境变化发生的代谢功能改变的有效手段[34]。在本试验中,湖羊在传染病(细菌)、免疫疾病、能量代谢以及折叠、分类和降解等生物代谢通路的富集高于卡拉库尔羊;而卡拉库尔羊则在核苷酸代谢、复制和修复、氨基酸代谢、碳水化合物代谢、膜运输等生物代谢通路的富集更高。其中微生物的复制和修复、核苷酸代谢通路被证明与细菌增长密切相关[35]。本试验中卡拉库尔羊瘤胃菌群的多样性和丰富度均高于湖羊组,说明卡拉库尔羊良好的适应性可能与其瘤胃微生物的复制和修复、核苷酸代谢通路的表达有关。李勇[36]研究表明,瘤胃内许多微生物能够共享代谢通路,瘤胃微生物的差异性并不会改变瘤胃微生物的功能。因此,造成瘤胃菌群代谢途径差异的原因尚需要更加深入的研究。
4 结论湖羊与卡拉库尔羊瘤胃菌群结构和瘤胃微生物的代谢通路具有一定的相似性,但卡拉库尔羊瘤胃菌群的多样性和丰富度均高于湖羊,一些特异性菌属相对丰度和生物代谢通路富集的差异可能与卡拉库尔羊适应高粗饲料饲粮的特性有关。
| [1] |
韩亚儒. 中国绵羊地方品种的种质特性及其生态分布规律的研究[D]. 硕士学位论文. 泰安: 山东农业大学, 2016. HAN Y R. The research of Chinese indigenous sheep breeds germplasm characteristics and their ecological distributing law[D]. Master's Thesis. Tai'an: Shandong Agricultural University, 2016. (in Chinese) |
| [2] |
托合塔森·皮达巴依. 新疆地方绵羊品种遗传资源保护与利用[J]. 新疆畜牧业, 2016(增刊1): 2-4. PIDABAYI T H T S. Protection and utilization of genetic resources of Xinjiang local sheep breeds[J]. Animal Husbandry in Xinjiang, 2016(Suppl.1): 2-4 (in Chinese). |
| [3] |
邹声雷, 张子荣, 卢海燕, 等. 新疆引进湖羊繁育指标调查报告[J]. 现代畜牧兽医, 2020(5): 44-48. ZOU S L, ZHANG Z R, LU H Y, et al. Recent survey on breeding and reproduction index of Hu-sheep being introduced to Xinjiang[J]. Modern Journal of Animal Husbandry and Veterinary Medicine, 2020(5): 44-48 (in Chinese). |
| [4] |
安尧汶, 谢鹏贵, 沈虎泉, 等. 湖羊引种以及在新疆养殖经验[J]. 中国畜禽种业, 2018, 14(9): 35-36. AN Y W, XIE P G, SHEN H Q, et al. Introduction of Hu sheep and experience of breeding in Xinjiang[J]. The Chinese Livestock and Poultry Breeding, 2018, 14(9): 35-36 (in Chinese). DOI:10.3969/j.issn.1673-4556.2018.09.028 |
| [5] |
EDWARDS J E, FORSTER R J, CALLAGHAN T M, et al. PCR and Omics based techniques to study the diversity, ecology and biology of anaerobic fungi: insights, challenges and opportunities[J]. Frontiers in Microbiology, 2017, 8: 1657. DOI:10.3389/fmicb.2017.01657 |
| [6] |
INDUGU N, VECCHIARELLI B, BAKER L D, et al. Comparison of rumen bacterial communities in dairy herds of different production[J]. BMC Microbiology, 2017, 17(1): 190. DOI:10.1186/s12866-017-1098-z |
| [7] |
JEWELL K A, MCCORMICK C A, ODT C L, et al. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency[J]. Applied and Environmental Microbiology, 2015, 81(14): 4697-4710. DOI:10.1128/AEM.00720-15 |
| [8] |
SHABAT S K B, SASSON G, DORON-FAIGENBOIM A, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants[J]. The ISME Journal, 2016, 10(12): 2958-2972. DOI:10.1038/ismej.2016.62 |
| [9] |
HENDERSON G, COX F, GANESH S, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range[J]. Scientific Reports, 2015, 5: 14567. DOI:10.1038/srep14567 |
| [10] |
CUI X X, WANG Z F, YAN T H, et al. Rumen bacterial diversity of Tibetan sheep (Ovis aries) associated with different forage types on the Qinghai-Tibetan Plateau[J]. Canadian Journal of Microbiology, 2019, 65(12): 859-869. DOI:10.1139/cjm-2019-0154 |
| [11] |
KING E E, SMITH R P, ST-PIERRE B, et al. Differences in the rumen methanogen populations of lactating Jersey and Holstein dairy cows under the same diet regimen[J]. Applied and Environmental Microbiology, 2011, 77(16): 5682-5687. DOI:10.1128/AEM.05130-11 |
| [12] |
韩帅. 育肥方式对呼伦贝尔羊和绒山羊瘤胃发酵及瘤胃微生物数量的影响[D]. 硕士学术论文. 呼和浩特: 内蒙古农业大学, 2014. HAN S. Effect of fattening ways on rumen fermentation and the number of several rumen microorganisms in Hulun Buir sheep and cashmere goats[D]. Master's Thesis. Hohhot: Inner Mongolia Agricultural University, 2014. (in Chinese) |
| [13] |
中华人民共和国农业部. NY/T 816-2004: 肉羊饲养标准[S]. 北京: 中国农业出版社, 2004. The People's Republic of China. NY/T 816-2004: meat sheep feeding standard[S]. Beijing: China Agriculture Press, 2004. (in Chinese) |
| [14] |
刘开朗, 卜登攀, 王加启, 等. 六个不同品种牛的瘤胃微生物群落的比较分析[J]. 中国农业大学学报, 2009, 14(1): 13-18. LIU K L, BU D P, WANG J Q, et al. Comparative analysis of rumen microbial communities in six species of cattle[J]. Journal of China Agricultural University, 2009, 14(1): 13-18 (in Chinese). DOI:10.3321/j.issn:1007-4333.2009.01.003 |
| [15] |
CHANG J J, YAO X T, ZUO C X, et al. The gut bacterial diversity of sheep associated with different breeds in Qinghai province[J]. BMC Veterinary Research, 2020, 16(1): 254. DOI:10.1186/s12917-020-02477-2 |
| [16] |
HUANG J Q, LI Y J, LUO Y Z. Bacterial community in the rumen of Tibetan sheep and Gansu alpine fine-wool sheep grazing on the Qinghai-Tibetan Plateau, China[J]. The Journal of General and Applied Microbiology, 2017, 63(2): 122-130. DOI:10.2323/jgam.2016.08.003 |
| [17] |
普宣宣. 饲粮NFC/NDF对卡拉库尔羊消化、瘤胃发酵及瘤胃微生物的影响[D]. 硕士学术论文. 阿拉尔: 塔里木大学, 2020. PU X X. Effects of dietary NFC/NDF ratio on digestion, rumen fermentation and the rumen microorganism in Karakul sheep[D]. Master's Thesis. Alaer: Tarim University, 2020. (in Chinese) |
| [18] |
范文斌, 李长青, 高仙灵, 等. 放牧蒙古羊瘤胃细菌随季节变化规律的研究[J]. 中国畜牧兽医, 2019, 46(8): 2315-2321. FAN W B, LI C Q, GAO X L, et al. Study on the seasonal variation of rumen bacteria in grazing Mongolian sheep[J]. China Animal Husbandry & Veterinary Medicine, 2019, 46(8): 2315-2321 (in Chinese). |
| [19] |
李与琦, 阳建华, 张涛, 等. 日粮中添加不同比例微贮棉秆对湖羊瘤胃微生物区系的影响[J]. 微生物学报, 2020, 60(8): 1592-1604. LI Y Q, YANG J H, ZHANG T, et al. Effect of different proportions fermented cotton stalk intake on rumen microflora of lambs[J]. Acta Microbiologica Sinica, 2020, 60(8): 1592-1604 (in Chinese). |
| [20] |
陈芸, 刘旗, 邓俊良, 等. 基于MiSeq分析川中黑山羊瘤胃细菌的多样性及群落结构[J]. 湖南农业大学学报(自然科学版), 2017, 43(3): 286-291. CHEN Y, LIU Q, DENG J L, et al. Analysis the bacterial diversity in rumen and their community structure from Chuanzhong black goat using MiSeq sequencing technology[J]. Journal of Hunan Agricultural University (Natural Sciences), 2017, 43(3): 286-291 (in Chinese). |
| [21] |
WONGWILAIWALIN S, LAOTHANACHAREON T, MHUANTONG W, et al. Comparative metagenomic analysis of microcosm structures and lignocellulolytic enzyme systems of symbiotic biomass-degrading consortia[J]. Applied Microbiology and Biotechnology, 2013, 97(20): 8941-8954. DOI:10.1007/s00253-013-4699-y |
| [22] |
MYER P R, SMITH T P L, WELLS J E, et al. Rumen microbiome from steers differing in feed efficiency[J]. PLoS One, 2015, 10(6): e0129174. DOI:10.1371/journal.pone.0129174 |
| [23] |
JAMI E, WHITE B A, MIZRAHI I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency[J]. PLoS One, 2014, 9(1): e85423. DOI:10.1371/journal.pone.0085423 |
| [24] |
FERNANDO S C, PURVIS H T 2ND, NAJAR F Z, et al. Rumen microbial population dynamics during adaptation to a high-grain diet[J]. Applied and Environmental Microbiology, 2010, 76(22): 7482-7490. DOI:10.1128/AEM.00388-10 |
| [25] |
WILLEMS A, COLLINS M D. Phylogenetic placement of Dialister pneumosintes (formerly Bacteroides pneumosintes) within the Sporomusa subbranch of the Clostridium subphylum of the gram-positive bacteria[J]. International Journal of Systematic Bacteriology, 1995, 45(2): 403-405. DOI:10.1099/00207713-45-2-403 |
| [26] |
马力, 徐世晓, 刘宏金, 等. 不同物候期牧草对放牧牦牛瘤胃内环境参数及瘤胃微生物多样性的影响[J]. 动物营养学报, 2019, 31(2): 681-691. MA L, XU S X, LIU H J, et al. Effects of forage grass in different phenological periods on ruminal environment parameters and rumen microbial diversity of free-range yak (Bos grunniens)[J]. Chinese Journal of Animal Nutrition, 2019, 31(2): 681-691 (in Chinese). DOI:10.3969/j.issn.1006-267x.2019.02.024 |
| [27] |
邢磊, 赵圣国, 郑楠, 等. 对以纤维素为唯一碳源条件下瘤胃细菌群落动态变化的研究[J]. 中国畜牧兽医, 2019, 46(8): 2322-2329. XING L, ZHAO S G, ZHENG N, et al. The research of dynamic changes of rumen bacteria community cultured in the medium with cellulose as the sole carbon source[J]. China Animal Husbandry & Veterinary Medicine, 2019, 46(8): 2322-2329 (in Chinese). |
| [28] |
MICHALET-DOREAU B, FERNANDEZ I, PEYRON C, et al. Fibrolytic activities and cellulolytic bacterial community structure in the solid and liquid phases of rumen contents[J]. Reproduction, Nutrition, Development, 2001, 41(2): 187-194. DOI:10.1051/rnd:2001122 |
| [29] |
FONDEVILA M, DEHORITY B A. Interactions between Fibrobacter succinogenes, Prevotella ruminicola, and Ruminococcus flavefaciens in the digestion of cellulose from forages[J]. Journal of Animal Science, 1996, 74(3): 678-684. DOI:10.2527/1996.743678x |
| [30] |
TAJIMA K, AMINOV R I, NAGAMINE T, et al. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR[J]. Applied and Environmental Microbiology, 2001, 67(6): 2766-2774. DOI:10.1128/AEM.67.6.2766-2774.2001 |
| [31] |
NYONYO T, SHINKAI T, MITSUMORI M. Improved culturability of cellulolytic rumen bacteria and phylogenetic diversity of culturable cellulolytic and xylanolytic bacteria newly isolated from the bovine rumen[J]. FEMS Microbiology Ecology, 2014, 88(3): 528-537. DOI:10.1111/1574-6941.12318 |
| [32] |
KOIKE S, YOSHITANI S, KOBAYASHI Y, et al. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria[J]. FEMS Microbiology Letters, 2003, 229(1): 23-30. DOI:10.1016/S0378-1097(03)00760-2 |
| [33] |
李子健. 不同生理阶段奶牛瘤胃细菌菌群数量与多样性的比较研究[D]. 硕士学位论文. 呼和浩特: 内蒙古农业大学, 2018. LI Z J. Comparative study on rumen bacteria quantity and diversity of dairy cows in different physiological phases[D]. Master's Thesis. Hohhot: Inner Mongolia Agricultural University, 2018. (in Chinese) |
| [34] |
李偲奇. 不同料型日粮对育肥羊生产性能、胃肠道微生物组和代谢组的影响研究[D]. 硕士学位论文. 泰安: 山东农业大学, 2020. LI S Q. Effects of diets types on performance, gastrointestinal microbiome and metabolome of fattening lambs[D]. Master's Thesis. Taian: Shandong Agricultural University, 2020. (in Chinese) |
| [35] |
王莹, 张建海, 王勇亮, 等. 包装方式和贮藏温度对羊肉微生物数量、细菌多样性和代谢途径的影响[J/OL]. 食品科学: 1-13. (2021-11-19)[2021-12-01]. http://kns.cnki.net/kcms/detail/11.2206.TS.20211118.1057.014.html. WANG Y, ZHANG J H, WANG Y L, et al. Effect of packaging method and storage temperature on microbial quantity, bacterial diversity and metabolic pathways in mutton[J/OL]. Food Science: 1-13. (2021-11-19)[2021-12-01]. http://kns.cnki.net/kcms/detail/11.2206.TS.20211118.1057.014.html. (in Chinese) |
| [36] |
李勇. 两种不同物理形态开食料对羔羊生长和瘤胃发育的影响及其机制研究[D]. 博士学位论文. 兰州: 甘肃农业大学, 2020. LI Y. Effects of starter feeds of two different physical forms on growth and rument development of lambs and its mechanisms[D]. Ph. D. Thesis. Lanzhou: Gansu Agricultural University, 2020. (in Chinese) |




