2. 甘肃省农业科学院畜草与绿色农业研究所, 甘肃省牛羊种质与秸秆饲料化重点实验室, 兰州 730070
2. Key Laboratory for Sheep, Goat and Cattle Germplasm and Straw Feed in Gansu Province, Institute of Livestock Grass and Green Agriculture, Gansu Academy of Agricultural Sciences, Lanzhou 730070, China
人类胃肠道栖居着1013~1014个的微生物,其在宿主免疫调节、营养代谢、维持肠道屏障结构完整性和抵御病原体方面发挥着重要作用[1-2]。胃肠道微生物群失调不仅与肠内疾病有关,如肠屏障功能障碍、炎症性肠病和结直肠癌;还与多种肠外疾病有关,包括糖尿病、神经系统疾病、关节炎、肝癌和严重急性呼吸系统综合征冠状病毒2(SARS-CoV-2)感染[3-8]。因此,维持胃肠道微生物群的稳态对人类及动物的健康是非常重要的,并有可能改变预防和治疗疾病的策略。早在20世纪初,人们就认识到了保持微生物区系健康的重要性,人们发现食用酸奶和特定的细菌混合物与延长寿命和预防疾病密切相关。益生菌最初被定义为“一种活的微生物,当给予足够的量时,会给宿主带来健康益处”[9-11]。近年来,随着“健康养殖”理念的提出,胃肠道微生物群的稳态越发受到人们的关注。抗生素曾被用来治疗疾病和促进动物生长,但其导致的耐抗生素细菌的出现和肉类中抗生素残留问题是一大弊端[12-16],且抗生素具有杀死致病菌和益生菌的双重作用,会扰乱胃肠道微生物群稳态,促进致病菌定植,从而导致疾病[17-19]。肠道菌群失衡和抗生素的滥用会激活宿主肠道的免疫反应、炎症反应和促进胃肠道蠕动,对宿主健康不利[20-21]。更严重的是,幼龄动物断奶前滥用抗生素,会导致幼龄动物胃肠道微生物菌群发育不成熟,对饲粮营养成分的吸收和胃肠道屏障系统的发育造成永久性的负面影响[22-23]。益生菌与宿主的互作是个复杂的过程,其中之一的关键点是胃肠道黏膜屏障,但益生菌对胃肠道黏膜屏障的作用机制尚不明晰。因此,本文总结了近年来益生菌对胃肠道黏膜屏障调控的相关研究进展,通过益生菌对胃肠道上皮屏障的调控、对胃肠道黏液层的调控和对炎症反应的调控等方面进行论述,以期为益生菌对胃肠道黏膜屏障的调控以及维持胃肠道微生物群稳态提供一定的理论依据。
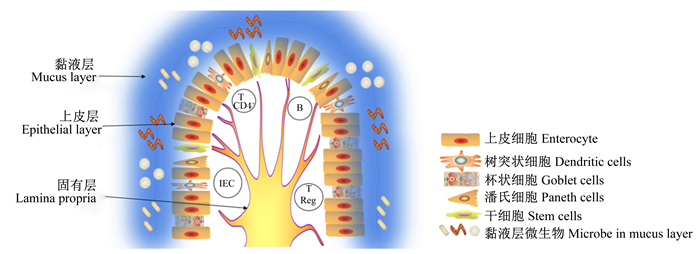
1 肠道黏膜屏障与免疫系统胃肠道是机体与外界环境接触的最大界面,与免疫系统之间联系密切[24-25]。微生物在这种联系中起关键作用,菌群组成调控宿主免疫系统,反过来宿主免疫系统影响菌群组成。宿主免疫系统调控有益微生物的组成,控制特定细菌的过度生长,同时通过肠道黏膜屏障系统对致病菌的入侵和定植做出相应的反应[26]。免疫系统与病原体之间的相互作用也受微生物的调控,微生物可以直接与致病菌互作或间接刺激免疫系统对致病菌做出相关的反应。因此,当免疫系统在共生、互惠和致病菌这三者之间建立一个适当的平衡时,肠道内环境的稳态得以维持[27]。肠道黏膜屏障是由一系列不均匀的实体组成的复杂生态系统,其由细胞外部分和细胞部分构成[28]。细胞外部分主要是杯状细胞分泌的黏液组成的黏液层,其中还包含一系列微生物。黏液的存在对促进食物通过、防止消化酶对上皮细胞的作用和防止致病菌定植至关重要[29]。细胞部分主要由肠道上皮细胞和下面的固有层组成。肠道上皮细胞包含5种不同类型的细胞:干细胞、潘氏细胞、可吸收上皮细胞、杯状细胞和肠内分泌细胞[30]。固有层由树突状细胞、上皮内树突状细胞、巨噬细胞、上皮内淋巴细胞、T调节细胞、T CD4+淋巴细胞、B淋巴细胞和浆细胞组成[31]。其中,巨噬细胞定位上皮细胞下区域,专门吞噬潜在有害微生物,并清除凋亡细胞和细胞碎片。树突状细胞在黏液层摄取致病菌并迁移到淋巴结,在淋巴结激活T细胞和炎症反应。浆细胞和T细胞通过免疫球蛋白、细胞因子和炎症介质调控肠道体液免疫[31-32](图 1)。在人类中,肠上皮细胞每3~5 d更新1次,这一机制本身具有免疫保护作用,其可以清除感染和受损伤的细胞[33]。此外,肠上皮细胞的通透性也具有免疫活性,受紧密连接的调控[34]。
|
T CD4+:T CD4+淋巴细胞T CD4+ lymphocytes;B:B淋巴细胞B lymphocytes;IEC:上皮内树突状细胞intraepithelial dendritic cell;T Reg:T调节细胞T regulatory cells。 图 1 肠黏膜屏障示意图 Fig. 1 Schematic diagram of the intestinal mucosal barrier |
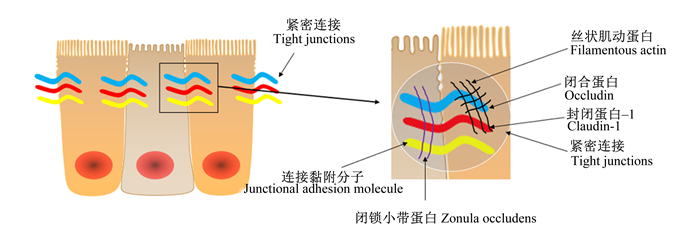
肠道上皮细胞的功能之一是在外部环境和免疫系统之间建立一个物理屏障,这个屏障的完整性和功能性是支持营养物质和有益分子与机体交换的关键,且可以阻止有害物质入侵。上皮细胞的选择性通透性是由紧密连接蛋白介导的,该蛋白主要负责上皮屏障功能[35]。在健康肠道中,紧密连接蛋白密封上皮细胞之间的间隙,形成一个物理屏障,防止促炎分子通过。当紧密连接受损时,有毒分子可以通过[34]。简单来说,紧密连接位于肠上皮细胞的顶端,它们由跨膜蛋白组成,胞外与邻近紧密连接的相似结构域相互作用,胞内与细胞骨架连接(图 2)。当紧密连接蛋白的表达或定位发生改变时,这种物理屏障的功能就会受到损害,可能会出现“肠漏”[36]。“肠漏”的特点是上皮细胞对分子或化合物的渗透性增加,这些分子或化合物从肠腔扩散到固有层。这种情况可以在体外通过跨上皮细胞电阻参数测量,或在体内通过肠通透性测试测量[37]。“肠漏”是多种病理状况的表现,如炎症性肠病、肠易激综合征和乳糜泻,所有特征都是持续的炎症和组织损伤[38]。
|
图 2 肠道上皮紧密连接示意图 Fig. 2 Schematic diagram of intestinal epithelial tight junctions |
饲粮中的多种化合物已经被证明可以调节紧密连接蛋白的表达[24]。类似地,益生菌也可以调节紧密连接蛋白的表达或定位(表 1),改变肠黏膜屏障功能,进而对致病菌的入侵做出反应。例如,无菌小鼠用尼氏大肠杆菌(Escherichia coli Nissle,EcN)或K12型大肠杆菌(MG1655)感染,分离小肠上皮细胞,分析闭锁小带蛋白-1(zonula occludens-1,ZO-1)和闭锁小带蛋白-2(zonula occludens-2,ZO-2)的基因和蛋白表达。结果表明,EcN定植无菌小鼠后,小鼠小肠上皮细胞中ZO-1的mRNA水平显著上调,但不会上调ZO-2的mRNA水平。因此,EcN可以通过上调ZO-1的mRNA水平保护黏膜的通透性[39];用极化的T84上皮细胞单层作为模型监测肠致病性大肠杆菌(enteropathogenic Escherichia coli,EPEC)E2348/69菌株感染后屏障的破坏。EPEC与EcN共孵育或在EPEC感染后加入EcN消除了屏障破坏,使用DNA微阵列(芯片)技术分析与EcN孵育的T84细胞,发现了300多个基因的表达改变。EcN改变了ZO-2蛋白的表达,ZO-2的表达随时间的推移向细胞边界重新分布而增强肠道黏膜屏障。这项研究表明,EcN是一种重要的诱导信号,导致破坏的上皮屏障的恢复[40]。肠出血性大肠杆菌(enterohemorrhagic Escherichia coli,EHEC)O157 ∶ H7感染可导致细胞间紧密连接中断,导致急性腹泻、出血性结肠炎等症状,用鼠李糖乳杆菌GG(Lactobacillus rhamnosus GG)预处理极化的MDCK-Ⅰ和T84细胞可以减少EHEC O157 ∶ H7感染诱导的形态学变化,且保护了上皮单分子层免受EHEC诱导的封闭蛋白-1(claudin-1)和ZO-1紧密连接蛋白的分配(用益生菌处理的上皮细胞单层比单独感染病原菌的单层细胞拥有更高水平的ZO-1表达,且可以防止大肠杆菌O157 ∶ H7引起的claudin-1再分配的变化),进而预防EHEC诱导的破坏[41]。干酪乳杆菌(Lactobacillus casei)DN-114001对致病菌株的黏附没有抑制作用。但干酪乳杆菌DN-114001抑制EPEC诱导的细胞旁通透性的增加(108 CFU/mL的干酪乳杆菌显著抑制EPEC诱导的改变,跨膜电阻抗降低和ZO-1再分配)[42]。细胞单层暴露于嗜热链球菌(Streptococcus thermophilus)ATCC19258或嗜酸乳杆菌(Lactobacillus acidophilus)ATCC4356活体,显著抑制了EIEC诱导的黏附、侵袭和生理功能障碍,且维持ZO-1、增强α-辅肌动蛋白(α-actinin)和闭合蛋白(occludin)的磷酸化水平[43]。在T84细胞系中,婴儿双歧杆菌(Bifidobacterium infantis)降低了封闭蛋白-2(claudin-2)的表达,提高了ZO-1和occludin的表达;用婴儿双歧杆菌的条件培养基处理T84细胞,封闭蛋白-4(claudin-4)、ZO-1和occludin蛋白表达增强,而claudin-2蛋白表达随着时间的推移而降低[44]。结肠腺癌细胞(Caco-2)经植物乳杆菌(Lactobacillus plantarum)MB452处理后,4种紧密连接蛋白(occludin、ZO-1、ZO-2和cingulin)的荧光强度均高于未处理的对照组[45]。健康受试者在十二指肠内注射植物乳杆菌WCFS1或安慰剂(llacebo)6 h后,在紧密连接结构附近,ZO-1和occludin表达明显增加[38]。
|
|
表 1 调节紧密连接的益生菌列表 Table 1 List of probiotics modulating tight junctions |
胃肠道上皮被一层黏液所覆盖,黏液是一种覆盖肠道上皮的流变性物质,是抵御各种有害分子、微生物感染和不同腔内环境的保护屏障[46]。健康的黏液层在预防炎症和感染性疾病中起着重要作用。胃肠道中的黏液是由肠道上皮中的杯状细胞产生的,主要是黏蛋白(MUC),MUC是一种高分子量的糖蛋白,主要有2种形式:分泌型黏蛋白(MUC2、MUC5AC、MUC5B和MUC6),负责黏液层的形成;跨膜黏蛋白(MUC1、MUC4、MUC13、MUC16),其功能目前尚不清楚,但可能参与信号通路[47-49]。特异性黏液蛋白表达的改变与消化道疾病如克罗恩病[50]和溃疡性结肠炎相关[51],这凸显了这些蛋白在胃肠道中的重要性。
3.2 益生菌调控胃肠道黏液层某些益生菌已被证明可以调节MUC的表达,从而影响黏液层的性质,并间接调节肠道免疫系统(表 2)。植物乳杆菌299v与HT-29细胞孵育后MUC2和MUC3的mRNA表达量提高[52]。用尼氏大肠杆菌与HT-29细胞共同培养,可导致MUC2、MUC3、MUC5AC和MUC5A的mRNA表达量提高[53]。在肠上皮细胞单层表面添加干酪乳杆菌GG可显著提高MUC2的表达,且随着干酪乳杆菌GG添加量的升高,MUC2蛋白密度显著增加[54]。在动物模型的体内研究中,大白鼠每天口服益生菌混合物VSL#3,连续7 d;治疗后,基底腔黏液含量增加60%。此外,将分离的大鼠结肠环暴露于VSL#3益生菌配方中,显著刺激了结肠黏蛋白的分泌和MUC2基因的表达;而MUC1和MUC3基因表达仅略有升高。体外研究表明,培养细胞与VSL#3细菌共同培养没有显示黏液分泌增加。然而,条件培养基中所含的细菌分泌产物显著刺激了MUC的分泌效果。在VSL#3所含的3种细菌群(乳酸杆菌、双歧杆菌和链球菌)中,乳酸杆菌是体外黏液分泌最强的促进剂[55]。鱼饲料中添加腐败希瓦氏菌(Shewanella putrefaciens),结果表明,MUC和免疫球蛋白T重链(IGHT)表达上调[56]。罗伊氏乳杆菌E(Lactobacillus reuteri E)与猪胃黏蛋白预培养后,再与HT-29细胞系共培养,HT-29细胞系中MUC2、MUC5AC和白细胞介素-10(IL-10)表达显著上调[57]。
|
|
表 2 益生菌对黏蛋白的调控 Table 2 Regulation of mucins by probiotics |
先天免疫系统是抵御感染或组织损伤的第1道防线,先天免疫的细胞和分子在遇到微生物或其他“危险信号”时迅速激活[58]。这些细胞通过使用模式识别受体如Toll样受体(Toll-like receptors,TLRs)和核苷酸结合寡聚化结构域样受体(nucleotide-binding and oligomerization domain,NLRs)来响应病原体相关的分子模式(pathogen-associated molecular pattern,PAMPs)和损伤相关的分子模式(damage associated molecular patterns,DAMPs)[59-61]。作为NLRs家族的一员,炎症小体是一组大型的细胞质多聚蛋白复合物,可加工和切割促炎细胞因子,如白细胞介素-1β(IL-1β)和白细胞介素-18(IL-18)前体,使之成熟[62-67]。成熟后的IL-1β和IL-18向免疫系统发出危险警报,并增强促炎免疫反应[68]。一般来说,炎症小体复合物由传感器、适配器和效应器组成。以NLRP3炎性小体为例,由传感器NLRP3、适配器凋亡相关斑点样蛋白(ASC)和效应器半胱天冬酶-1(Caspase-1)组成。作为最典型的炎症小体,NLRP3参与了许多不同的疾病病理过程[69]。NLRP3的功能对健康状态很重要,例如它有助于葡聚糖硫酸钠(dextran sulfate sodium,DSS)诱导的结肠炎小鼠肠道组织损伤的恢复[70]。然而,NLRP3的过度激活会导致一些炎症性疾病,包括脓毒症休克[71]、Ⅱ型糖尿病[72]、类风湿性关节炎[73]和阿尔茨海默氏病[74]。
4.2 益生菌对NLRP3的调控NLRP3的活化被抑制,可降低促炎细胞因子的产生,促炎细胞因子减少在一定程度上可缓解炎症进程。某些微生物可以通过抑制炎症小体组装(活化)减少促炎细胞因子的产生,从而达到缓解炎症进程的效果。例如粪肠杆菌(Enterococcus faecalis),热杀死的粪肠杆菌处理THP-1细胞系可抑制Caspase-1活化和减少IL-1β的产生,进而缓解炎症进程。小鼠体内试验中粪肠杆菌可改善肠道损伤严重程度[75]。Caspase-1作为NLRP3的效应器,是NLRP3完整组装不可缺少的部分,Caspase-1的活化被抑制使得NLRP3的组装(激活)不能完成,导致IL-1β前体的成熟被抑制,进一步使得IL-1β表达量减少。IL-1β是一种促炎细胞因子,其表达量减少可改善炎症的严重程度。在高脂饮食诱导的非酒精性脂肪肝大鼠肝脏脂肪变性和炎症反应中,乳酸双歧杆菌(Bifidobacterium lactis)V9可抑制NLRP3和ASC的mRNA表达,且可以抑制促炎细胞因子[如白细胞介素-6(IL-6)、IL-1β和肿瘤坏死因子-α(TNF-α)]的产生[76]。ASC作为NLRP3小体的适配器,是NLRP3小体完整组装的重要部分,ASC表达量的减少使得NLRP3小体的组装被抑制,导致促炎细胞因子前体的成熟被抑制,进一步使得促炎细因子表达量降低,从而缓解炎症进程。约氏杆菌L531预处理能抑制婴儿沙门氏菌诱导的小肠NLRC4和NLRP3炎症小体的活化,从而抑制炎症反应[77]。炎症小体的组装(活化)被抑制,导致促炎细胞因子前体的活化被抑制,进而降低促炎细胞因子的释放,从而缓解炎症进程。用婴儿双歧杆菌(Bifidobacterium infantis)30治疗DSS诱导的结肠炎,结果表明,婴儿双歧杆菌显著降低结肠组织中NLRP3的mRNA水平,显著下调结肠组织中促炎细胞因子TNF-α和IL-1β的mRNA水平,显著提高结肠组织中occludin-1、occludin和claudin-1紧密连接的mRNA水平[78]。NLRP3 mRNA表达水平降低,说明NLRP3小体的组装(激活)被抑制,导致促炎细胞因子前体的活化被抑制,进而降低促炎细胞因子的表达量,从而缓解炎症进程。此外还有干酪乳杆菌,在多发性硬化症中,用干酪乳杆菌(1×109 CFU/mL)进行治疗,可显著降低炎症小体基因的表达[79]。炎症小体基因表达量降低,说明炎症小体的组装(激活)被抑制,导致促炎细胞因子前体的活化被抑制,从而降低促炎细胞因子的表达量,进而缓解炎症进程。
5 小结宿主免疫系统与肠道微生物之间存在一定的联系,微生物可以影响宿主免疫系统的状态,反过来宿主免疫系统对微生物的存在形式也有影响。了解二者间的互作机制对宿主的健康生长和抵抗疾病方面具有深远的影响,也是未来畜牧业健康持续发展的理论前提。益生菌可调控宿主胃肠道上皮紧密连和黏液层,并且对炎症小体也有调控作用。宿主胃肠道上皮紧密连接的改变会影响胃肠道上皮的通透性,进而导致宿主对营养物质的吸收和对致病菌的抵抗发生改变。黏液层的改变影响致病菌的定植。炎症小体的组装(激活)与宿主体内的炎症进程密切相关,益生菌具有调控炎症小体的能力,进而使得炎症进程发生改变,炎症进程对宿主的免疫系统至关重要。尽管国内外科研工作者对微生物与宿主互作的研究已经取得了丰硕的成果,但关于微生物具体是怎样调节宿主免疫系统的机制尚不精确,例如微生物本身还是其代谢产物对宿主免疫系统起主要调节作用,这种调节作用的信号是如何被宿主免疫系统识别并做出相应的反馈,这些问题还需要进一步研究。相信随着科技的发展,人们将逐步完善微生物与机体免疫系统之间的互作这一复杂过程的相关理论研究,为微生物促进机体的健康提供新的思路。
| [1] |
GENTILE C L, WEIR T L. The gut microbiota at the intersection of diet and human health[J]. Science, 2018, 362(6416): 776-780. DOI:10.1126/science.aau5812 |
| [2] |
RINNINELLA E, RAOUL P, CINTONI M, et al. What is the healthy gut microbiota composition?A changing ecosystem across age, environment, diet, and diseases[J]. Microorganisms, 2019, 7(1): 14. DOI:10.3390/microorganisms7010014 |
| [3] |
LI X, ATKINSON M A. The role for gut permeability in the pathogenesis of type 1 diabetes—a solid or leaky concept?[J]. Pediatric Diabetes, 2015, 16(7): 485-492. DOI:10.1111/pedi.12305 |
| [4] |
IMHANN F, VICH VILA A, BONDER M J, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease[J]. Gut, 2018, 67(1): 108-119. DOI:10.1136/gutjnl-2016-312135 |
| [5] |
TILG H, ADOLPH T E, GERNER R R, et al. The intestinal microbiota in colorectal cancer[J]. Cancer Cell, 2018, 33(6): 954-964. DOI:10.1016/j.ccell.2018.03.004 |
| [6] |
MUÑOZ L, BORRERO M J, ÚBEDA M, et al. Intestinal immune dysregulation driven by dysbiosis promotes barrier disruption and bacterial translocation in rats with cirrhosis[J]. Hepatology, 2019, 70(3): 925-938. DOI:10.1002/hep.30349 |
| [7] |
PONZIANI F R, BHOORI S, CASTELLI C, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease[J]. Hepatology, 2019, 69(1): 107-120. DOI:10.1002/hep.30036 |
| [8] |
ZUO T, ZHANG F, LUI G C Y, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization[J]. Gastroenterology, 2020, 159(3): 944-955.e8. DOI:10.1053/j.gastro.2020.05.048 |
| [9] |
BROWN A C, VALIERE A. Probiotics and medical nutrition therapy[J]. Nutrition in Clinical Care, 2004, 7(2): 56-68. |
| [10] |
RHODES J M. Colonic mucus and ulcerative colitis[J]. Gut, 1997, 40(6): 807-808. DOI:10.1136/gut.40.6.807 |
| [11] |
WANG H Y, LEE I S, BRAUN C, et al. Effect of probiotics on central nervous system functions in animals and humans: a systematic review[J]. Journal of Neurogastroenterology and Motility, 2016, 22(4): 589-605. DOI:10.5056/jnm16018 |
| [12] |
BROWN K, UWIERA R R E, KALMOKOFF M L, et al. Antimicrobial growth promoter use in livestock: a requirement to understand their modes of action to develop effective alternatives[J]. International Journal of Antimicrobial Agents, 2017, 49(1): 12-24. DOI:10.1016/j.ijantimicag.2016.08.006 |
| [13] |
CHO I, YAMANISHI S, COX L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity[J]. Nature, 2012, 488(7413): 621-626. DOI:10.1038/nature11400 |
| [14] |
LAXMINARAYAN R, DUSE A, WATTAL C, et al. Antibiotic resistance-the need for global solutions[J]. The Lancet.Infectious Diseases, 2013, 13(12): 1057-1098. DOI:10.1016/S1473-3099(13)70318-9 |
| [15] |
EARNSHAW S, MENDEZ A, MONNET D L, et al. Global collaboration to encourage prudent antibiotic use[J]. The Lancet.Infectious Diseases, 2013, 13(12): 1003-1004. DOI:10.1016/S1473-3099(13)70315-3 |
| [16] |
ALLEN H K, LEVINE U Y, LOOFT T, et al. Treatment, promotion, commotion: antibiotic alternatives in food-producing animals[J]. Trends in Microbiology, 2013, 21(3): 114-119. DOI:10.1016/j.tim.2012.11.001 |
| [17] |
RASHID M U, WEINTRAUB A, NORD C E. Effect of new antimicrobial agents on the ecological balance of human microflora[J]. Anaerobe, 2012, 18(2): 249-253. DOI:10.1016/j.anaerobe.2011.11.005 |
| [18] |
BUFFIE C G, PAMER E G. Microbiota-mediated colonization resistance against intestinal pathogens[J]. Nature Reviews Immunology, 2013, 13(11): 790-801. DOI:10.1038/nri3535 |
| [19] |
UBEDA C, PAMER E G. Antibiotics, microbiota, and immune defense[J]. Trends in Immunology, 2012, 33(9): 459-466. DOI:10.1016/j.it.2012.05.003 |
| [20] |
SPEES A M, LOPEZ C A, KINGSBURY D D, et al. Colonization resistance: battle of the bugs or Ménage à Trois with the host?[J]. PLoS Pathogens, 2013, 9(11): e1003730. DOI:10.1371/journal.ppat.1003730 |
| [21] |
RIVERA-CHÁVEZ F, ZHANG L F, FABER F, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella[J]. Cell Host & Microbe, 2016, 19(4): 443-454. |
| [22] |
JI S K, JIANG T, YAN H, et al. Ecological restoration of antibiotic-disturbed gastrointestinal microbiota in foregut and hindgut of cows[J]. Frontiers in Cellular and Infection Microbiology, 2018, 8: 79. DOI:10.3389/fcimb.2018.00079 |
| [23] |
WOLIN M J. Fermentation in the rumen and human large intestine[J]. Science, 1981, 213(4515): 1463-1468. DOI:10.1126/science.7280665 |
| [24] |
DE SANTIS S, CAVALCANTI E, MASTRONARDI M, et al. Nutritional keys for intestinal barrier modulation[J]. Frontiers in Immunology, 2015, 6: 612. |
| [25] |
FARHADI A, BANAN A, FIELDS J, et al. Intestinal barrier: an interface between health and disease[J]. Journal of Gastroenterology and Hepatology, 2003, 18(5): 479-497. DOI:10.1046/j.1440-1746.2003.03032.x |
| [26] |
LLORENTE C, SCHNABL B. The gut microbiota and liver disease[J]. Cellular and Molecular Gastroenterology and Hepatology, 2015, 1(3): 275-284. DOI:10.1016/j.jcmgh.2015.04.003 |
| [27] |
PETERSON C T, SHARMA V, ELMÉN L, et al. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota[J]. Clinical and Experimental Immunology, 2015, 179(3): 363-377. DOI:10.1111/cei.12474 |
| [28] |
DEMEO M T, MUTLU E A, KESHAVARZIAN A, et al. Intestinal permeation and gastrointestinal disease[J]. Journal of Clinical Gastroenterology, 2002, 34(4): 385-396. DOI:10.1097/00004836-200204000-00003 |
| [29] |
HARRISON O J, MALOY K J. Innate immune activation in intestinal homeostasis[J]. Journal of Innate Immunity, 2011, 3(6): 585-593. DOI:10.1159/000330913 |
| [30] |
KRAEHENBUHL J P, PRINGAULT E, NEUTRA M R. Review article: intestinal epithelia and barrier functions[J]. Alimentary Pharmacology & Therapeutics, 1997, 11(Suppl 3): 3-9. |
| [31] |
SCHENK M, MUELLER C. The mucosal immune system at the gastrointestinal barrier[J]. Best Practice & Research: Clinical Gastroenterology, 2008, 22(3): 391-409. |
| [32] |
KANAI T, ILYAMA R, ISHIKURA T, et al. Role of the innate immune system in the development of chronic colitis[J]. Journal of Gastroenterology, 2002, 37(Suppl 14): 38-42. |
| [33] |
BISCHOFF S C, BARBARA G, BUURMAN W, et al. Intestinal permeability-a new target for disease prevention and therapy[J]. BMC Gastroenterology, 2014, 14(1): 189. |
| [34] |
SUZUKI T. Regulation of intestinal epithelial permeability by tight junctions[J]. Cellular and Molecular Life Sciences, 2013, 70(4): 631-659. |
| [35] |
FARQUHAR M G, PALADE G E. Junctional complexes in various epithelia[J]. Journal of Cell Biology, 1963, 17(2): 375-412. DOI:10.1083/jcb.17.2.375 |
| [36] |
ULLUWISHEWA D, ANDERSON R C, MCNABB W C, et al. Regulation of tight junction permeability by intestinal bacteria and dietary components[J]. The Journal of Nutrition, 2011, 141(5): 769-776. DOI:10.3945/jn.110.135657 |
| [37] |
MÉNARD S, CERF-BENSUSSAN N, HEYMAN M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens[J]. Mucosal Immunology, 2010, 3(3): 247-259. DOI:10.1038/mi.2010.5 |
| [38] |
KARCZEWSKI J, TROOST F J, KONINGS I, et al. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier[J]. American Journal of Physiology: Gastrointestinal and Liver Physiology, 2010, 298(6): G851-G859. DOI:10.1152/ajpgi.00327.2009 |
| [39] |
UKENA S N, SINGH A, DRINGENBERG U, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity[J]. PLoS One, 2007, 2(12): e1308. DOI:10.1371/journal.pone.0001308 |
| [40] |
ZYREK A A, CICHON C, HELMS S, et al. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair[J]. Cellular Microbiology, 2007, 9(3): 804-816. DOI:10.1111/j.1462-5822.2006.00836.x |
| [41] |
JOHNSON-HENRY K C, DONATO K A, SHEN-TU G, et al. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157 ∶ H7-induced changes in epithelial barrier function[J]. Infection and Immunity, 2008, 76(4): 1340-1348. DOI:10.1128/IAI.00778-07 |
| [42] |
PARASSOL N, FREITAS M, THOREUX K, et al. Lactobacillus casei DN-114 001 inhibits the increase in paracellular permeability of enteropathogenic Escherichia coli-infected T84 cells[J]. Research in Microbiology, 2005, 156(2): 256-262. DOI:10.1016/j.resmic.2004.09.013 |
| [43] |
RESTA-LENERT S, BARRETT K E. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC)[J]. Gut, 2003, 52(7): 988-997. DOI:10.1136/gut.52.7.988 |
| [44] |
EWASCHUK J B, DIAZ H, MEDDINGS L, et al. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function[J]. American Journal of Physiology: Gastrointestinal and Liver Physiology, 2008, 295(5): G1025-G1034. DOI:10.1152/ajpgi.90227.2008 |
| [45] |
ANDERSON R C, COOKSON A L, MCNABB W C, et al. Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation[J]. BMC Microbiology, 2010, 10: 316. DOI:10.1186/1471-2180-10-316 |
| [46] |
KEBOUCHI M, HAFEEZ Z, LE ROUX Y, et al. Importance of digestive mucus and mucins for designing new functional food ingredients[J]. Food Research International, 2020, 131: 108906. DOI:10.1016/j.foodres.2019.108906 |
| [47] |
CORFIELD A P, MYERSCOUGH N, LONGMAN R, et al. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease[J]. Gut, 2000, 47(4): 589-594. DOI:10.1136/gut.47.4.589 |
| [48] |
CORNICK S, TAWIAH A, CHADEE K. Roles and regulation of the mucus barrier in the gut[J]. Tissue Barriers, 2015, 3(1/2): e982426. |
| [49] |
MACK D R, GAGINELLA T S, SHERMAN P M. Effect of colonic inflammation on mucin inhibition of Escherichia coli RDEC-1 binding in vitro[J]. Gastroenterology, 1992, 102(4 Pt 1): 1199-1211. |
| [50] |
BUISINE M P, DESREUMAUX P, LETEURTRE E, et al. Mucin gene expression in intestinal epithelial cells in Crohn's disease[J]. Gut, 2001, 49(4): 544-551. DOI:10.1136/gut.49.4.544 |
| [51] |
GAJENDRAN M, LOGANATHAN P, JIMENEZ G, et al. A comprehensive review and update on ulcerative colitis[J]. Disease-a-Month, 2019, 65(12): 100851. DOI:10.1016/j.disamonth.2019.02.004 |
| [52] |
MACK D R, MICHAIL S, WEI S, et al. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression[J]. The American Journal of Physiology, 1999, 276(4): G941-G950. |
| [53] |
HAFEZ M M. Upregulation of intestinal mucin expression by the probiotic bacterium E. coli Nissle 1917[J]. Probiotics and Antimicrobial Proteins, 2012, 4(2): 67-77. DOI:10.1007/s12602-012-9092-0 |
| [54] |
MATTAR A F, TEITELBAUM D H, DRONGOWSKI R A, et al. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model[J]. Pediatric Surgery International, 2002, 18(7): 586-590. DOI:10.1007/s00383-002-0855-7 |
| [55] |
CABALLERO-FRANCO C, KELLER K, DE SIMONE C, et al. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells[J]. American Journal of Physiology: Gastrointestinal and Liver Physiology, 2007, 292(1): G315-G322. DOI:10.1152/ajpgi.00265.2006 |
| [56] |
CHEN Z C, CEBALLOS-FRANCISCO D, GUARDIOLA F A, et al. Influence of skin wounds on the intestinal inflammatory response and barrier function: protective role of dietary Shewanella putrefaciens SpPdp11 administration to gilthead seabream (Sparus aurata L.)[J]. Fish & Shellfish Immunology, 2020, 99: 414-423. |
| [57] |
DUDÍK B, KIÑOVÁ SEPOVÁ H, BILKA F, et al. Mucin pre-cultivated Lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells[J]. Antonie van Leeuwenhoek, 2020, 113(8): 1191-1200. DOI:10.1007/s10482-020-01426-1 |
| [58] |
HATO T, DAGHER P C. How the innate immune system senses trouble and causes trouble[J]. Clinical Journal of the American Society of Nephrology, 2015, 10(8): 1459-1469. DOI:10.2215/CJN.04680514 |
| [59] |
O'NEILL L A J, GOLENBOCK D, BOWIE A G. The history of Toll-like receptors-redefining innate immunity[J]. Nature Reviews.Immunology, 2013, 13(6): 453-460. DOI:10.1038/nri3446 |
| [60] |
KAWAI T, AKIRA S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature Immunology, 2010, 11(5): 373-384. DOI:10.1038/ni.1863 |
| [61] |
KAWAI T, AKIRA S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650. DOI:10.1016/j.immuni.2011.05.006 |
| [62] |
LAMKANFI M, DIXIT V M. Mechanisms and functions of inflammasomes[J]. Cell, 2014, 157(5): 1013-1022. DOI:10.1016/j.cell.2014.04.007 |
| [63] |
HENAO-MEJIA J, ELINAV E, THAISS C A, et al. Inflammasomes and metabolic disease[J]. Annual Review of Physiology, 2014, 76: 57-78. DOI:10.1146/annurev-physiol-021113-170324 |
| [64] |
RATHINAM V A K, VANAJA S K, FITZGERALD K A. Regulation of inflammasome signaling[J]. Nature Immunology, 2012, 13(4): 333-342. DOI:10.1038/ni.2237 |
| [65] |
LAMKANFI M, DIXIT V M. Inflammasomes and their roles in health and disease[J]. Annual Review of Cell and Developmental Biology, 2012, 28: 137-161. DOI:10.1146/annurev-cellbio-101011-155745 |
| [66] |
SWANSON K V, DENG M, TING J P Y. The NLRP3 inflammasome: molecular activation and regulation to therapeutics[J]. Nature Reviews Immunology, 2019, 19(8): 477-489. |
| [67] |
MANGAN M S J, OLHAVA E J, ROUSH W R, et al. Targeting the NLRP3 inflammasome in inflammatory diseases[J]. Nature Reviews: Drug Discovery, 2018, 17(8): 588-606. |
| [68] |
SCHRODER K, TSCHOPP J. The Inflammasomes[J]. Cell, 2010, 140(6): 821-832. |
| [69] |
LU F F, LAN Z X, XIN Z Q, et al. Emerging insights into molecular mechanisms underlying pyroptosis and functions of inflammasomes in diseases[J]. Journal of Cellular Physiology, 2020, 235(4): 3207-3221. |
| [70] |
ALLEN I C, TEKIPPE E M, WOODFORD R M T, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer[J]. Journal of Experimental Medicine, 2010, 207(5): 1045-1056. |
| [71] |
LEE S, NAKAHIRA K, DALLI J, et al. NLRP3 inflammasome deficiency protects against microbial sepsis via increased lipoxin B4 synthesis[J]. American Journal of Respiratory and Critical Care Medicine, 2017, 196(6): 713-726. |
| [72] |
LEE H M, KIM J J, KIM H J, et al. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes[J]. Diabetes, 2013, 62(1): 194-204. |
| [73] |
MATHEWS R J, ROBINSON J I, BATTELLINO M, et al. Evidence of NLRP3-inflammasome activation in rheumatoid arthritis (RA); genetic variants within the NLRP3-inflammasome complex in relation to susceptibility to RA and response to anti-TNF treatment[J]. Annals of the Rheumatic Diseases, 2014, 73(6): 1202-1210. |
| [74] |
HENEKA M T, KUMMER M P, STUTZ A, et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice[J]. Nature, 2013, 493(7434): 674-678. |
| [75] |
CHUNG I C, OUYANG C N, YUAN S N, et al. Pretreatment with a heat-killed probiotic modulates the NLRP3 inflammasome and attenuates colitis-associated colorectal cancer in mice[J]. Nutrients, 2019, 11(3): 516. |
| [76] |
YAN Y, LIU C Y, ZHAO S M, et al. Probiotic Bifidobacterium lactis V9 attenuates hepatic steatosis and inflammation in rats with non-alcoholic fatty liver disease[J]. AMB Express, 2020, 10(1): 101. |
| [77] |
XIA B, YU J, HE T, et al. Lactobacillus johnsonii L531 ameliorates enteritis via elimination of damaged mitochondria and suppression of SQSTM1-dependent mitophagy in a Salmonella infantis model of piglet diarrhea[J]. FASEB Journal, 2020, 34(2): 2821-2839. |
| [78] |
SHENG K L, HE S M, SUN M, et al. Synbiotic supplementation containing Bifidobacterium infantis and xylooligosaccharides alleviates dextran sulfate sodium-induced ulcerative colitis[J]. Food & Function, 2020, 11(5): 3964-3974. |
| [79] |
DIGEHSARA S G, NAME N, SARTIPNIA N, et al. Analysis of inflammasomes and CYP27B1 genes in cuprizone demyelinated C57BL/6 mice and evaluation of Th1 and Th2 patterns after oral administration of Lactobacillus casei strain T2 (IBRC-M10783)[J]. Microbial Pathogenesis, 2021, 155: 104931. |




