2. 青海大学畜牧兽医科学院, 西宁 810016;
3. 青海东牧湾农牧科技开发有限公司, 海东 811600
2. Academy of Animal Husbandry and Veterinary Sciences, Qinghai University, Xining 810016, China;
3. Qinghai Dongmuwan Agriculture and Animal Husbandry Technology Development Co., Ltd., Haidong 811600, China
牦牛(Bos grunniens)是青藏高原及周边地区的优良畜种,为高原人民提供了最基本的生产和生活资料。牦牛与牧民长期以来的相互依存关系,使得牦牛已成为高原历史文化的重要组成部分,牦牛也为亚洲高原经济发展做出了巨大贡献[1]。近年来,过度放牧导致了草场的严重退化,牦牛冷季存活率和出栏率严重下降[2]。因此,牦牛的营养管理,特别是冷季传统饲养方式的转变,对于提高牦牛的健康、营养水平和生长发育尤为重要。
瘤胃作为将光合作用吸收的光能转化为供人类食用的奶、肉等易消化化合物之间的重要中间媒介[3],在反刍动物生长发育中发挥着重要的功能。瘤胃中存在细菌、古菌、真菌和原生动物等复杂的微生物群落,能够将难以消化的植物转化为供家畜维持生长所需的营养物质和能量[4]。细菌约占瘤胃微生物总量的95%[5]。瘤胃的正常活动与瘤胃微生物发酵关系密切,是反刍动物维持正常生理活动的必要保证[6]。以往研究表明,奶牛、肉牛、牦牛、肉羊和山羊等反刍动物瘤胃微生物群落组成对饲粮粗蛋白质、粗纤维、能量水平或饲养方式极其敏感[1, 7-9]。据报道,高精料饲粮能促进瘤胃上皮细胞的增殖、分化、转化生长因子-β1(TGF-β1)和转录因子过氧化物酶体增殖激活受体(PPAR-α)的表达,进而促进瘤胃上皮乳头的发育[10]。此外,瘤胃代谢产物被微生物用于自我增殖,并与控制微生物代谢及相关营养途径的代谢因子相互作用[11]。16S rRNA高通量测序技术是分析微生物群落多样性的可靠方法,已被广泛应用于动物和人类胃肠道微生物的研究[12-13]。据韩旭峰[14]报道,瘤胃微生物受到饲粮营养水平的影响。因此,为更好地解释瘤胃微生物是如何受饲养方式转变的影响,进而如何改变瘤胃发育,对促进牦牛瘤胃发育,提高牦牛生长性能和养殖效率具有重要意义。
鉴于此,本研究从青藏高原高寒牧区牦牛生产实际出发,系统研究放牧和舍饲2种饲养方式对青海大通牦牛瘤胃组织结构及瘤胃菌群多样性和组成的影响,提出适宜大通牦牛的冷季饲养方式,为了解牦牛瘤胃内特定微生物功能和提高牦牛瘤胃发育水平提供理论依据。
1 材料与方法 1.1 试验设计本试验采用配对试验设计原则,选取30头体重[(213.50±13.12) kg]相近、健康状况良好的2岁大通公牦牛,随机分为放牧组和舍饲组,每组15头牦牛。放牧组牦牛每天07:00—19:00进行放牧,放牧期间自由采食和饮水。舍饲组牦牛分3个独立圈舍,每个圈舍随机分配5头,每日饲喂全混合日粮(TMR),TMR配方参见文献[15]。每日分2次(08:00和17:00)饲喂,自由采食和饮水。试验预试期15 d,正试期190 d。
1.2 样品采集与测定 1.2.1 瘤胃测序样本的采集试验结束后,将24头试验牦牛全部禁食24 h,断水8 h后进行屠宰,每头牦牛采集50 mL瘤胃液,混匀后过滤,取其中20 mL瘤胃液封装至5个5 mL冻存管后立刻放入液氮罐,运回实验室于-80 ℃保存,以备DNA提取和高通量测序分析。
1.2.2 瘤胃组织切片样品的采集与测定为观察瘤胃组织形态发育情况,每头牦牛剪取2 cm×2 cm大小的瘤胃组织3份,经4%多聚甲醛固定后作为固定组织标本。所有标本送样至成都里来生物科技有限公司按病理检验标准操作程序(SOP)进行。标本经脱水、修剪、包埋、切片、染色、封片等,最后镜检。采用麦克奥迪实业集团有限公司生产的BA200Digital数码三目摄像显微摄像系统对切片进行镜检图像采集,每张切片于40倍下观察全部组织,选择要测量的区域并采集40倍和400倍图片待测。用数码三目摄像显微镜(BA410Digital,麦克奥迪实业集团有限公司)观察瘤胃组织,用Motic Images Advanced软件测量瘤胃乳头长度、乳头宽度、上皮厚度、角质层厚度和肌层厚度[16]。
1.2.3 瘤胃细菌DNA抽提与PCR扩增根据E.Z.N.A.® soil DNA kit(Omega Bio-Tek, Norcross,美国)说明书进行24个牦牛瘤胃细菌群落总DNA抽提,使用1%的琼脂糖凝胶电泳检测DNA的提取质量,使用NanoDrop2000测定DNA浓度和纯度;使用引物338F(5′-ACTCCTACGGGAGGCAGCAG-3′)和806R(5′-GGACTACHVGGGTWTCTAAT-3′)对16S rRNA基因V3~V4可变区进行PCR扩增(PCR仪:ABI GeneAmp® 9700型)。扩增程序如下:95 ℃预变性3 min;95 ℃变性30 s,55 ℃退火30 s, 72 ℃延伸30 s, 27个循环;72 ℃稳定延伸10 min,最后在4 ℃进行保存。PCR反应体系为20 μL:5×TransStart FastPfu缓冲液4 μL,2.5 mmol/L dNTPs 2 μL,上游引物(5 μmol/L)0.8 μL,下游引物(5 μmol/L)0.8 μL,TransStart FastPfu DNA聚合酶0.4 μL,模板DNA 10 ng,添加双蒸水至20 μL。每个样本3个重复[17]。
1.2.4 瘤胃细菌Illumina MiSeq测序根据上海美吉生物医药科技有限公司的标准方案,使用Illumina公司的Illumina MiSeq平台对配对末端进行测序,然后将纯化的扩增子等摩尔汇集在一起。
将同一样本的PCR产物混合后使用2%琼脂糖凝胶回收PCR产物,利用AxyPrep DNA Gel Extraction Kit(Axygen Biosciences,美国)进行回收产物纯化,2%琼脂糖凝胶电泳检测,并用QuantusTM Fluorometer(Promega, 美国)对回收产物进行定量[18]。使用NEXTFLEX® Rapid DNA-Seq Kit进行建库[18]。利用Illumina公司的Illumina MiSeq平台进行测序,由上海美吉生物医药科技有限公司完成。
1.2.5 测序数据处理使用fastp[18](https://github.com/OpenGene/fastp,version 0.20.0)软件对原始测序序列进行质控,使用FLASH[19](http://www.cbcb.umd.edu/software/flash,version 1.2.7)软件进行拼接。使用UPARSE[18]软件(http://drive5.com/uparse/,version 7.1),根据97%[20]的相似度对序列进行操作分类单元(OTU)聚类并剔除嵌合体。利用RDP classifier[21](http://rdp.cme.msu.edu/,version 2.2)对每条序列进行物种分类注释,比对Silva 16S rRNA数据库(v138),设置比对阈值为70%,对每个OTU代表性序列进行分类分析。
1.3 统计分析数据采用Excel 2013进行整理,采用SPSS 19.0软件对放牧组与舍饲组大通牦牛试验数据采用独立样本t检验进行分析比较,结果用平均值±标准差来表示,以P>0.05表示组间差异不显著,P<0.05表示组间差异显著,P<0.01表示组间差异极显著。
2 结果 2.1 饲养方式对大通牦牛瘤胃组织结构的影响由图 1和表 1可知,饲养方式显著或极显著影响了大通牦牛的瘤胃组织结构指标(P<0.05或P<0.01)。舍饲组牦牛瘤胃乳头长度、乳头宽度、上皮厚度和角质层厚度显著或极显著高于放牧组牦牛(P<0.05或P<0.01),瘤胃肌层厚度显著低于放牧组牦牛(P<0.05)。
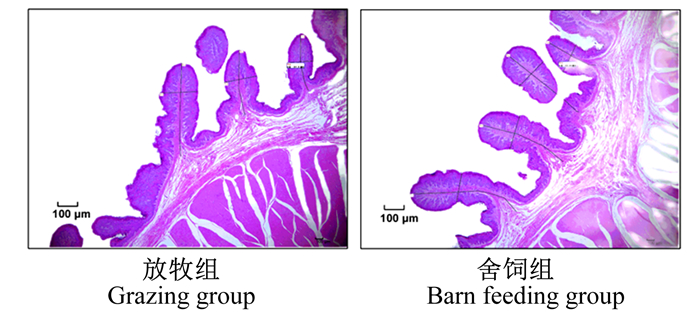
|
图 1 大通牦牛瘤胃组织形态学观察 Fig. 1 Observation of rumen histomorphology of Datong yaks |
|
|
表 1 饲养方式对大通牦牛瘤胃组织结构的影响 Table 1 Effects of feeding pattern on rumen tissue structure of Datong yaks |
由表 2可知,饲养方式显著或极显著影响了大通牦牛瘤胃菌群Alpha多样性(P<0.05或P<0.01)。放牧组牦牛瘤胃菌群Chao1指数、ACE指数和Shannon指数极显著高于舍饲组牦牛(P<0.01),Simpson指数则显著低于舍饲组牦牛(P<0.05)。
|
|
表 2 饲养方式对大通牦牛瘤胃菌群Alpha多样性的影响 Table 2 Effects of feeding pattern on Alpha diversity of rumen bacterial community of Datong yaks |
图 2的主成分分析(PCA)显示,受不同饲养方式的影响,牦牛瘤胃菌群Beta多样性存在差异,表现为放牧组牦牛瘤胃菌群Beta多样性极显著高于舍饲组牦牛(P<0.01)。
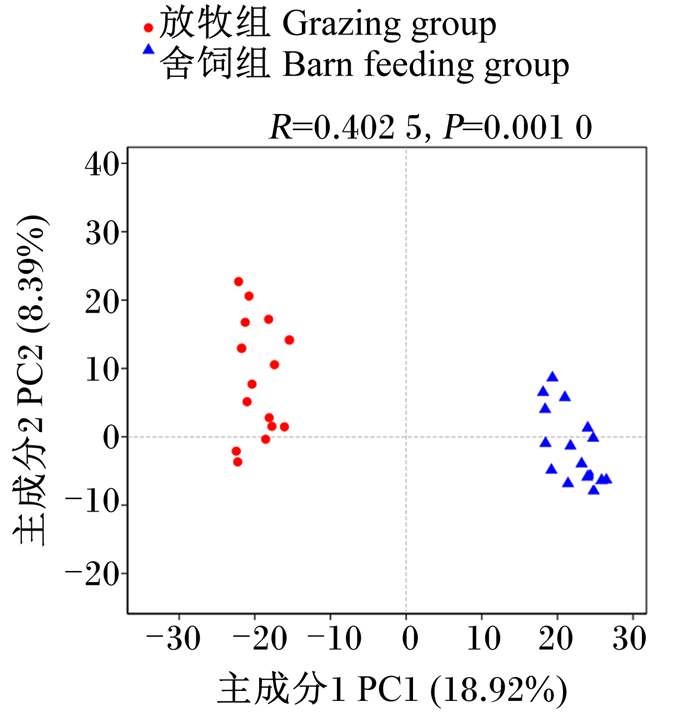
|
图 2 大通牦牛瘤胃菌群PCA Fig. 2 PCA of rumen bacterial community of Datong yaks |
由图 3和表 3可知,门水平上,舍饲组牦牛瘤胃菌群中厚壁菌门(Firmicutes)、变形菌门(Proteobacteria)和蓝细菌门(Cyanobacteria)的相对丰度显著高于放牧组牦牛(P<0.05),拟杆菌门(Bacteroidetes)和放线菌门(Actinobacteria)的相对丰度显著或极显著低于放牧组牦牛(P<0.05或P<0.01)。
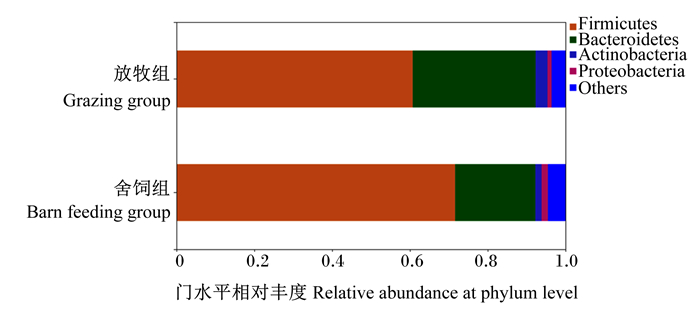
|
Firmicutes:厚壁菌门;Bacteroidetes:拟杆菌门;Actinobacteria:放线菌门;Proteobacteria:变形菌门;Others:其他。 图 3 大通牦牛瘤胃菌群门水平物种组成 Fig. 3 Species composition at phylum level of rumen bacterial community of Datong yaks |
|
|
表 3 大通牦牛瘤胃菌群中相对丰度在门水平>0.1%的物种 Table 3 Species relative abundance > 0.1% at phylum level in rumen bacterial community of Datong yaks |
由图 4和表 4可知,属水平上,舍饲组牦牛瘤胃菌群中克里斯滕森菌科_R-7群(Christensenellaceae_R-7_group)、瘤胃球菌属_2(Ruminococcus_2)、毛螺菌科_NK3A20群(Lachnospiraceae_NK3A20_group)、CAG-35、瘤胃球菌科_UCG-014(Ruminococcaceae_UCG-014)、瘤胃球菌科_UCG-001(Ruminococcaceae_UCG-001)、Candidatus_Saccharimonas的相对丰度显著或极显著高于放牧组牦牛(P<0.05或P<0.01),瘤胃球菌科_NK4A214群(Ruminococcaceae_NK4A214_group)、理研菌科_RC9肠道群(Rikenellaceae_RC9_gut_group)和普雷沃氏菌科_UCG-003(Prevotellaceae_UCG-003)的相对丰度显著或极显著低于放牧组牦牛(P<0.05或P<0.01)。
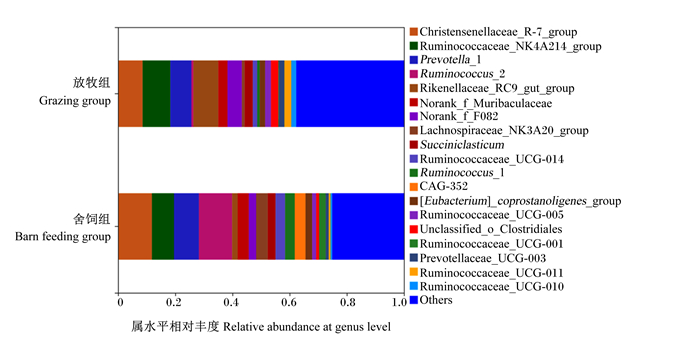
|
Christensenellaceae_R-7_group:克里斯滕森菌科_R-7群;Ruminococcaceae_NK4A214_group:瘤胃球菌科_NK4A214群;Prevotella_1:普氏菌属_1;Ruminococcus_2:瘤胃球菌属_2;Rikenellaceae_RC9_gut_group:理研菌科_RC9肠道群;Lachnospiraceae_NK3A20_group:毛螺菌科_NK3A20群;Succiniclasticum:解琥珀酸菌属;Ruminococcaceae_UCG-014:瘤胃球菌科_UCG-014;Ruminococcus_1:瘤胃球菌属_1;[Eubacterium]_coprostanoligenes_group:产粪甾醇真细菌群;Ruminococcaceae_UCG-005:瘤胃球菌科_UCG-005;Unclassified_o_Clostridiales:未分类梭菌目;Ruminococcaceae_UCG-001:瘤胃球菌科_UCG-001;Prevotellaceae_UCG-003:普雷沃氏菌科_UCG-003;Ruminococcaceae_UCG-011:瘤胃球菌科_UCG-011;Ruminococcaceae_UCG-010:瘤胃球菌科_UCG-010;Others:其他。 图 4 大通牦牛瘤胃菌群数水平物种组成 Fig. 4 Species composition at genus level of rumen bacterial community of Datong yaks |
|
|
表 4 大通牦牛瘤胃菌群中相对丰度在属水平>1%的物种 Table 4 Species relative abundance > 1% at genus level in rumen bacterial community of Datong yaks |
通过皮尔逊相关性研究瘤胃组织结构指标与属水平相对丰度差异较大的瘤胃菌群的关系(图 5)可知,Christensenellaceae_R-7_group和Ruminococcus_2的相对丰度与乳头长度和乳头宽度均呈极显著正相关(P<0.01),与上皮厚度和角质层厚度均呈显著正相关(P<0.05);Lachnospiraceae_NK3A20_group的相对丰度与乳头长度和乳头宽度均呈显著正相关(P<0.05),与肌层厚度呈显著负相关(P<0.05);Prevotellaceae_UCG-003的相对丰度与乳头长度和乳头宽度均呈显著负相关(P<0.05);Rikenellaceae_RC9_gut_group的相对丰度与乳头长度、乳头宽度、上皮厚度和角质层厚度均呈显著负相关(P<0.05)。
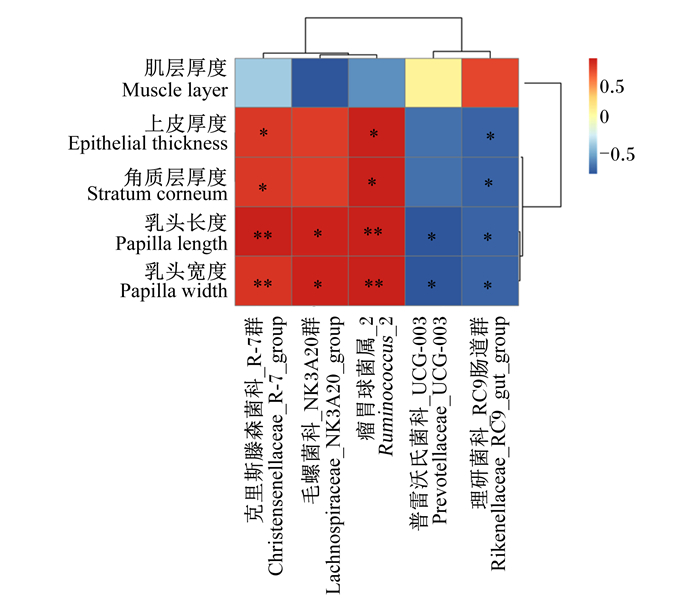
|
蓝色和红色分别代表负相关和正相关,* *表示极显著相关(P<0.01),*表示显著相关(P<0.05)。 The negative and positive correlations were represented by blue and red, respectively. * * mean extremely significant correlation (P < 0.01), and * mean significant correlation (P < 0.05). 图 5 大通牦牛瘤胃组织结构指标与瘤胃菌群的相关性分析 Fig. 5 Correlation analysis between rumen tissue structure indicators and rumen bacteria community of Datong yaks |
牦牛由于生长环境高寒低氧的特殊性,造成了牦牛对粗饲料具有较强的消化能力。瘤胃作为反刍动物消化粗饲料的大型厌氧发酵罐,通过瘤胃微生物的降解作用或发酵作用可将植物饲料转化为挥发性脂肪酸(VFA),为机体提供能量。据Sosin-Bzducha等[22]报道,瘤胃组织发育状况会显著影响反刍动物对饲料的消化利用程度,在犊牛饲粮中,用青贮玉米籽粒代替干玉米饲喂犊牛显著增加了瘤胃乳头长度,明显改善了瘤胃上皮厚度和角质层厚度。此外,瘤胃上皮对VFA的吸收速率在很大程度上取决于转运体的可用性和乳头表面积,有50%~85%的VFA被瘤胃上皮直接吸收,因而瘤胃的发育水平对瘤胃上皮细胞对VFA的吸收非常关键[23]。Miller等[23]研究表明,奶牛的瘤胃上皮细胞随着饲粮和生理变化而发生变化,从干奶牛饲粮到泌乳牛饲粮的转变显著促进了奶牛瘤胃乳头的发育,增强了瘤胃上皮细胞对VFA的吸收能力和适应性。本研究通过对表征瘤胃组织形态学特征等发育指标的评价,分析了牦牛瘤胃组织结构,以揭示饲养方式转变对大通牦牛瘤胃组织发育的影响。研究发现,舍饲组大通牦牛瘤胃乳头长度、乳头宽度、上皮厚度和角质层厚度均显著或极显著高于放牧组牦牛,可能是因为放牧组牦牛长期摄入的天然牧草数量有限,造成瘤胃中VFA的含量较低,一定程度上限制了瘤胃乳头、上皮和角质层的发育。Merchen等[24]报道了瘤胃中VFA的含量对羔羊瘤胃乳头、上皮和角质层的发育有显著影响,饲喂粗饲料会显著降低羔羊瘤胃乳头长度、乳头宽度、上皮厚度和角质层厚度,这与本研究结果一致。舍饲条件下,瘤胃吸收VFA的能力较强,瘤胃乳头发育较好,角质层的完整性以及角质层细胞的角化程度在很大程度上影响了瘤胃上皮对营养物质的吸收和运输,因此,舍饲组牦牛瘤胃上皮厚度和角质层厚度的增加促进了牦牛瘤胃的发育,促进了瘤胃对营养物质的吸收。本研究中,舍饲组牦牛瘤胃肌层厚度极显著低于放牧组,这可能是放牧组牦牛采食的是粗饲料的原因,研究表明,粗饲料可以刺激瘤胃壁肌肉的发育和提高肌层厚度[25]。
3.2 饲养方式对大通牦牛瘤胃菌群多样性和组成的影响本研究探讨了不同饲养方式对大通牦牛瘤胃菌群多样性和组成的影响。以往研究表明,饲养方式和饲粮能量水平会显著影响甘南牦牛的生长性能、瘤胃发酵参数和瘤胃菌群多样性与组成[26]。牦牛瘤胃核心菌群由10个不同的菌群组成,但其相对丰度可能存在差异[27]。本研究发现,饲养方式极显著影响了大通牦牛瘤胃菌群的Alpha多样性和Beta多样性,且均表现为放牧组牦牛极显著高于舍饲组牦牛。在门水平上,2种饲养方式均以Firmicutes、Proteobacteria、Bacteroidetes和Actinobacteria为大通牦牛的瘤胃优势菌群;另外,在属水平上,2种饲养方式间均以Christensenellaceae_R-7_group、Ruminococcaceae_NK4A214_group、Ruminococcus_2和Rikenellaceae_RC9_gut_group为大通牦牛的瘤胃优势菌群。Alpha多样性分析显示,放牧组牦牛和舍饲组牦牛瘤胃菌群多样性和丰富度各有不同,这与Zhou等[28]认为饲喂粗饲料牦牛瘤胃菌群多样性和丰富度与饲喂精饲料牦牛存在差异的研究结果一致,也与Fang等[29]的研究得出的放牧或室内养殖会对牦牛瘤胃菌群多样性造成不同的影响的结论一致。本研究结果说明,牦牛瘤胃菌群多样性和组成与饲养方式直接相关。
3.3 瘤胃组织结构指标与瘤胃菌群的相关性饲养方式对大通牦牛瘤胃菌群多样性和组成有显著影响。李宏等[30]研究表明,饲养水平对阿勒泰羊胃肠道发育、瘤胃发酵参数、消化酶活性及瘤胃微生物区系具有显著影响,较高的饲养水平能够显著改善阿勒泰羊胃肠道发育、促进瘤胃发酵和改善微生物区系。舍饲组牦牛瘤胃Christensenellaceae_R-7_group和Ruminococcus_2的相对丰度极显著高于放牧组牦牛,Lachnospiraceae_NK3A20_group和Rikenellaceae_RC9_gut_group相对丰度显著或极显著低于放牧组牦牛。Christensenellaceae_R-7_group是Firmicutes家族成员,研究表明,Christensenellaceae_R-7_group能够迅速对饲粮营养作出反映,参与胃肠道蛋白质分解代谢,与饲粮蛋白质水平呈正相关[31-32]。本研究中,舍饲组牦牛饲粮蛋白质水平高于放牧组牦牛,促进了瘤胃Christensenellaceae_R-7_group的定植,使其瘤胃中的相对丰度极显著高于放牧组牦牛。舍饲组牦牛瘤胃Ruminococcus_2的相对丰度极显著高于放牧组牦牛,Ruminococcus_2被认为是瘤胃中具有纤维降解能力的主要细菌,Ruminococcus_2相对丰度的改变可以从根本上改变瘤胃中营养物质的消化和利用[32]。Ruminococcus_2的生长与Toll样受体(Toll-like receptors,TLRs)基因表达呈正相关,TLRs基因能够触发免疫应答以维持宿主微生物的稳态,并能识别宿主细菌及其产物的诱导[33]。本研究中,Christensenellaceae_R-7_group的相对丰度与乳头长度和乳头宽度均呈极显著正相关,Ruminococcus_2的相对丰度与乳头长度、乳头宽度、上皮厚度和角质层厚度呈极显著正相关,这是由于舍饲组牦牛瘤胃发育较好,有利于营养物质的消化吸收,从而增加了Christensenellaceae_R-7_group和Ruminococcus_2的相对丰度,促进了瘤胃组织的发育。
此外,Lachnospiraceae_NK3A20_group的相对丰度与乳头长度和乳头宽度均呈显著正相关,与肌层厚度呈显著负相关。Lachnospiraceae_NK3A20_group是反刍动物胃肠道菌群的重要组成部分,与丁酸盐的产生密切相关。据报道,在去势牛的瘤胃中,丁酸可以显著增加瘤胃上皮乳头的表面积、长度和宽度[34]。本研究认为,Lachnospiraceae_NK3A20_group除了通过增加丁酸产量促进瘤胃的发育外,还能直接影响瘤胃的发育。随着Lachnospiraceae_NK3A20_group丰度的增加,牦牛瘤胃利用率显著提高,使牦牛在促进瘤胃生长发育的同时也获得了更多的能量。Rikenellaceae_RC9_gut_group的相对丰度与乳头长度、乳头宽度、上皮厚度和角质层厚度均呈显著负相关,Rikenellaceae_RC9_gut_group属于理研菌科(Rikenellaceae),在粗纤维的消化中起着重要作用[7]。舍饲组牦牛瘤胃Rikenellaceae_RC9_gut_group的相对丰度极显著低于放牧组牦牛,可能是舍饲组牦牛瘤胃中粗饲料含量较低的缘故。Prevotellaceae_UCG-003的相对丰度与乳头宽度呈显著负相关,研究表明,Prevotellaceae_UCG-003对瘤胃环境pH较为敏感,瘤胃pH越低,Prevotellaceae_UCG-003的相对丰度越低[35]。舍饲组牦牛由于瘤胃pH较低,因此Prevotellaceae_UCG-003的相对丰度也较低。研究发现Prevotellaceae_UCG-003可以提高饲料效率[36],同时Prevotellaceae_UCG-003还可以利用VFA参与糖代谢[37]。这说明舍饲组牦牛瘤胃中Prevotellaceae_UCG-003的减少促进了瘤胃的生长发育,但Prevotellaceae_UCG-003的减少促进乳头长度增加的机制仍有待进一步探索。
4 结论饲养方式显著或极显著影响了大通牦牛的瘤胃组织结构及瘤胃菌群多样性和组成,且瘤胃组织结构的变化与瘤胃菌群相关。本研究结果将为更好地了解牦牛瘤胃内特定菌群功能及其在促进瘤胃发育方面的相关机制提供理论依据。
| [1] |
LIANG C N, WANG L Z, WU X Y, et al. Genome-wide association study identifies loci for the polled phenotype in yak[J]. PLoS One, 2016, 11(7): e0158642. DOI:10.1371/journal.pone.0158642 |
| [2] |
MIAO F H, GUO Z G, XUE R, et al. Effects of grazing and precipitation on herbage biomass, herbage nutritive value, and yak performance in an alpine meadow on the Qinghai-Tibetan Plateau[J]. PLoS One, 2015, 10(6): e0127275. DOI:10.1371/journal.pone.0127275 |
| [3] |
JAMI E, WHITE B A, MIZRAHI I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency[J]. PLoS One, 2014, 9(1): e85423. DOI:10.1371/journal.pone.0085423 |
| [4] |
FLINT H J, BAYER E A, RINCON M T, et al. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis[J]. Nature Reviews Microbiology, 2008, 6(2): 121-131. DOI:10.1038/nrmicro1817 |
| [5] |
BRULC J M, ANTONOPOULOS D A, MILLER M E B, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(6): 1948-1953. DOI:10.1073/pnas.0806191105 |
| [6] |
DEUSCH S, CAMARINHA-SILVA A, CONRAD J, et al. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments[J]. Frontiers in Microbiology, 2017, 8: 1605. DOI:10.3389/fmicb.2017.01605 |
| [7] |
ZHANG J, SHI H T, WANG Y J, et al. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers[J]. Frontiers in Microbiology, 2017, 8: 2206. DOI:10.3389/fmicb.2017.02206 |
| [8] |
王书祥, 戴东文, 杨英魁, 等. 补饲精料对冷季放牧牦牛生长性能、瘤胃发酵及菌群结构的影响[J]. 动物营养学报, 2021, 33(11): 6266-6276. WANG S X, DAI D W, YANG Y K, et al. Effects of concentrate supplementation on growth performance, rumen fermentation and microbial community structure of grazing yaks in cold season[J]. Chinese Journal of Animal Nutrition, 2021, 33(11): 6266-6276 (in Chinese). DOI:10.3969/j.issn.1006-267x.2021.11.027 |
| [9] |
谢建林, 张巧娥, 张转弟, 等. 复合菌发酵稻草对安格斯牛生长性能、瘤胃发酵参数及菌群结构的影响[J]. 动物营养学报, 2021, 33(5): 2738-2751. XIE J L, ZHANG Q E, ZHANG Z D, et al. Effects of compound bacteria fermented rice straw on growth performance, rumen fermentation parameters and microbial structure of Angus cattle[J]. Chinese Journal of Animal Nutrition, 2021, 33(5): 2738-2751 (in Chinese). |
| [10] |
CONNOR E E, BALDWIN R L 6TH, WALKER M P, et al. Transcriptional regulators transforming growth factor-β1 and estrogen-related receptor-α identified as putative mediators of calf rumen epithelial tissue development and function during weaning[J]. Journal of Dairy Science, 2014, 97(7): 4193-4207. DOI:10.3168/jds.2013-7471 |
| [11] |
BANNINK A, VAN LINGEN H J, ELLIS J L, et al. The contribution of mathematical modeling to understanding dynamic aspects of rumen metabolism[J]. Frontiers in Microbiology, 2016, 7: 1820. |
| [12] |
LIN B, HENDERSON G, ZOU C X, et al. Characterization of the rumen microbial community composition of buffalo breeds consuming diets typical of dairy production systems in Southern China[J]. Animal Feed Science and Technology, 2015, 207: 75-84. DOI:10.1016/j.anifeedsci.2015.06.013 |
| [13] |
MCGOVERN E, WATERS S M, BLACKSHIELDS G, et al. Evaluating established methods for rumen 16S rRNA amplicon sequencing with mock microbial populations[J]. Frontiers in Microbiology, 2018, 9: 1365. DOI:10.3389/fmicb.2018.01365 |
| [14] |
韩旭峰. 日龄、日粮精粗比对陕北白绒山羊瘤胃微生物区系影响的研究[D]. 博士学位论文. 杨凌: 西北农林科技大学, 2015. HAN X F. Effects of age and dietary forage-to-concentrate ratios on rumen microbial flora of the Shaanbei white-cashmere goat[D]. Ph. D. Thesis. Yangling: Northwest A & F University, 2015. (in Chinese) |
| [15] |
姚喜喜, 王伟, 徐成体, 等. 放牧与舍饲对大通牦牛生长性能、瘤胃发酵参数、屠宰性能及肉品质的影响[J/OL]. 动物营养学报: 1-9. [2022-04-14]. http://kns.cnki.net/kcms/detail/11.5461.S.20220223.1103.022.html. YAO X X, WANG W, XU C T, et al. Effects of natural grazing and barn fattening on growth performance, rumen fermentation parameters, slaughter performance and meat quality of Datong yaks (Bos grunniens)[J/OL]. Chinese Journal of Animal Nutrition, 2022: 1-9. [2022-04-14]. http://kns.cnki.net/kcms/detail/11.5461.S.20220223.1103.022.html. (in Chinese) |
| [16] |
MALMUTHUGE N, LIANG G X, GUAN L L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes[J]. Genome Biology, 2019, 20(1): 172. DOI:10.1186/s13059-019-1786-0 |
| [17] |
WANG Q, GARRITY G M, TIEDJE J M, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy[J]. Applied and Environmental Microbiology, 2007, 73(16): 5261-5267. DOI:10.1128/AEM.00062-07 |
| [18] |
CHEN S F, ZHOU Y Q, CHEN Y R, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. DOI:10.1093/bioinformatics/bty560 |
| [19] |
MAGOČ T, SALZBERG S L. FLASH: fast length adjustment of short reads to improve genome assemblies[J]. Bioinformatics, 2011, 27(21): 2957-2963. DOI:10.1093/bioinformatics/btr507 |
| [20] |
EDGAR R C. UPARSE: highly accurate OTU sequences from microbial amplicon reads[J]. Nature Methods, 2013, 10(10): 996-998. DOI:10.1038/nmeth.2604 |
| [21] |
STACKEBRANDT E, GOEBEL B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology[J]. International Journal of Systematic and Evolutionary Microbiology, 1994, 44(4): 846-849. DOI:10.1099/00207713-44-4-846 |
| [22] |
SOSIN-BZDUCHA E, STRZETELSKI J, BOROWIEC F, et al. Effect of feeding ensiled maize grain on rumen development and calf rearing performance[J]. Journal of Animal & Feed Sciences, 2010, 19(2): 195-210. |
| [23] |
MILLER W F, TITGEMEYER E C, NAGARAJA T G, et al. Influence of cane molasses inclusion to dairy cow diets during the transition period on rumen epithelial development[J]. Animals, 2021, 11(5): 1230. DOI:10.3390/ani11051230 |
| [24] |
MERCHEN N R, BERGER L L. Effect of salinomycin level on nutrient digestibility and ruminal characteristics of sheep and feedlot performance of cattle[J]. Journal of Animal Science, 1985, 60(5): 1338-1346. DOI:10.2527/jas1985.6051338x |
| [25] |
PENNER G B, TANIGUCHI M, GUAN L L, et al. Effect of dietary forage to concentrate ratio on volatile fatty acid absorption and the expression of genes related to volatile fatty acid absorption and metabolism in ruminal tissue[J]. Journal of Dairy Science, 2009, 92(6): 2767-2781. DOI:10.3168/jds.2008-1716 |
| [26] |
张振宇, 梁春年, 姚喜喜, 等. 饲养方式和饲粮能量水平对牦牛生长性能、瘤胃发酵参数和瘤胃菌群的影响[J]. 动物营养学报, 2021, 33(6): 3343-3355. ZHANG Z Y, LIANG C N, YAO X X, et al. Effects of feeding model and dietary energy level on growth performance, rumen fermentation parameters and rumen bacterial community of yaks[J]. Chinese Journal of Animal Nutrition, 2021, 33(6): 3343-3355 (in Chinese). |
| [27] |
LIU C, WU H, LIU S J, et al. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type[J]. Frontiers in Microbiology, 2019, 10: 1116. DOI:10.3389/fmicb.2019.01116 |
| [28] |
ZHOU Z M, FANG L, MENG Q X, et al. Assessment of ruminal bacterial and archaeal community structure in yak (Bos grunniens)[J]. Frontiers in Microbiology, 2017, 8: 179. |
| [29] |
FANG L, ZHOU Z M, REN L P, et al. Ruminal bacterial diversity of yaks (Bos grunniens) fed by grazing or indoor regime on the Tibetan Plateau by analysis of 165 rRNA gene libraries[J]. Italian Journal of Animal Science, 2015, 14(4): 3970. DOI:10.4081/ijas.2015.3970 |
| [30] |
李宏, 宋淑珍, 高良霜, 等. 饲养水平对阿勒泰羊胃肠道发育、瘤胃发酵参数及瘤胃微生物区系的影响[J]. 草业学报, 2021, 30(4): 180-190. LI H, SONG S Z, GAO L S, et al. Effects of feeding level on the gastrointestinal development, rumen fermentation and rumen microbiota in Altay sheep[J]. Acta Prataculturae Sinica, 2021, 30(4): 180-190 (in Chinese). |
| [31] |
BONDER M J, TIGCHELAAR E F, CAI X H, et al. The influence of a short-term gluten-free diet on the human gut microbiome[J]. Genome Medicine, 2016, 8(1): 45. DOI:10.1186/s13073-016-0295-y |
| [32] |
KNISKERN M A, JOHNSTON C S. Protein dietary reference intakes may be inadequate for vegetarians if low amounts of animal protein are consumed[J]. Nutrition, 2011, 27(6): 727-730. DOI:10.1016/j.nut.2010.08.024 |
| [33] |
NATHANI N M, KOTHARI R K, PATEL A K, et al. Functional characterization reveals novel putative coding sequences in Prevotella ruminicola genome extracted from rumen metagenomic studies[J]. Journal of Molecular Microbiology and Biotechnology, 2015, 25(4): 292-299. DOI:10.1159/000437265 |
| [34] |
XU H, COLLINS J F, BAI L, et al. Epidermal growth factor regulation of rat NHE2 gene expression[J]. American Journal of Physiology.Cell Physiology, 2001, 281(2): C504-C513. DOI:10.1152/ajpcell.2001.281.2.C504 |
| [35] |
JIANG X, LIU X, LIU S, et al. Growth, rumen fermentation and plasma metabolites of Holstein male calves fed fermented corn gluten meal during the postweaning stage[J]. Animal Feed Science and Technology, 2019, 249: 1-9. DOI:10.1016/j.anifeedsci.2019.01.012 |
| [36] |
ZHAO X H, CHEN Z D, ZHOU S, et al. Effects of daidzein on performance, serum metabolites, nutrient digestibility, and fecal bacterial community in bull calves[J]. Animal Feed Science and Technology, 2017, 225: 87-96. DOI:10.1016/j.anifeedsci.2017.01.014 |
| [37] |
WARZECHA C M, COVERDALE J A, JANECKA J E, et al. Influence of short-term dietary starch inclusion on the equine cecal microbiome[J]. Journal of Animal Science, 2017, 95(11): 5077-5090. DOI:10.2527/jas2017.1754 |




