2. 浙江农业科学院省部共建农产品质量安全危害因子与风险防控国家重点实验室, 农产品质量安全与营养研究所, 杭州 310021;
3. 金华市农业科学研究院畜牧兽医研究所, 金华 321017
2. State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Agro-Product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China;
3. Institute of Animal Husbandry and Veterinary Medicine, Jinhua Academy of Agricultural Sciences, Jinhua 321017, China
肠道微生物群落是一个复杂的系统,存在于每个生命体中,以各种方式直接或间接地参与宿主的代谢过程,对宿主的营养、生理、免疫过程和生长性能起着重要的影响[1-3]。然而,当前对肠道微生物群落的研究大多是基于细菌群落,少有研究是针对真菌群落的[4-5]。真菌是肠道微生物的重要组成部分之一[6-8],越来越多的证据揭示了肠道真菌群落与宿主健康有着重要联系[9-12]。Köhler等[9]提出丝状真菌产生的次生代谢物(包括聚酮化合物、非核糖体肽、生物碱和萜烯等)具有调节机体代谢和药用的功能。此外,体外培养的威克汉姆酵母属(Wickerhamomyces)能够在4及25 ℃下分泌具有活性的胞外脂肪酶,可通过破坏甘油分子和脂肪酸之间的酯键来催化甘油三酯水解,促进脂质分解[10]。这些研究都揭示了肠道真菌参与机体的互作,除此之外,肠道真菌还可以通过调节肝肠轴缓解一系列的代谢疾病[11-12]。因此,肠道真菌也参与机体内部代谢及生命活动,其作用不可缺少。
金华猪是我国著名的地方品种,有“中国熊猫猪的美誉”,具有皮薄骨细、肉质鲜美、性成熟早、繁殖能力强和杂交优势显著的特点,反之,其具有生长速度慢、瘦肉率低、肌肉脂肪含量高等缺点[13-15]。鉴于肠道真菌在调节宿主生长代谢中的重要作用,由此推测,金华猪的回肠、结肠真菌结构可能与外来品种猪存在差异。然而,关于肠道真菌结构差异对脂质代谢的调控以及与背膘厚的相关性鲜有报道。因此,本试验选用不同肥瘦率的金华猪为研究对象,运用高通量测序技术探究其回肠、结肠的真菌结构,进一步探讨肠道真菌与脂肪沉积的相关性,从而为从微生物角度调控猪肉品质奠定理论基础。
1 材料与方法 1.1 试验设计与样品采集在金华市农业科学研究院试验猪场选择同批次的35头金华猪进行饲养,饲养期间自由采食和饮水,预防免疫和饲养管理按常规方法进行,饲粮组成及营养水平同我们前期的研究[16],具体见表 1。35头金华猪在270日龄称重后进行屠宰,测定其背膘厚、眼肌面积和瘦肉率,采集回肠、结肠中段内容物,并马上置于液氮速冻,然后干冰转运至实验室-80 ℃保存。选择瘦肉率最低(高脂肪组,H组)和瘦肉率最高(低脂肪组,L组)的5头猪用于肠道真菌结构分析。
|
|
表 1 基础饲粮组成及营养水平(风干基础) Table 1 Composition and nutrient levels of the basal diet (air-dry basis) |
使用DNA提取试剂盒(德国QIAGEN公司)提取回肠、结肠内容物DNA,并存储于-80 ℃。以提取的DNA为模板,使用ITS2引物ITS3-2024F(5’-GCATCGATGAAGAACGCAGC-3’)和ITS4-2409R(5’-TCCTCCGCTTATTGATATGC-3’)对真菌16S rRNA基因ITS2区域进行PCR扩增,扩增产物进行ITS测序分析[17]。
1.3 高通量测序及生物信息学分析测序由明科生物技术(杭州)有限公司完成,基于Illumina HiSeq 2500高通量测序平台对回肠、结肠内容物真菌ITS的ITS2区域进行扫描测序。采用QIIME软件(http://qiime.org/scripts/split_libraries_fastq.html)进行序列的质量控制及过滤,剔除低质量的DNA序列,获取高质量的DNA序列,然后在QIIME中调用uCLUST对优质序列按相似度97%进行聚类,定义为操作分类单元(operational taxonomic unit,OTU)并利用QIIME 2软件计算Alpha多样性指数[香农指数(Shannon index)、Chao1指数(Chao1 index)和辛普森指数(Simpson index)],使用Sliva和RPD数据库对所有OTU的代表性序列进行物种匹配,并对回肠、结肠内容物真菌群落的组成、多样性及测序深度进行表征,基于OTU聚类分析,绘制稀释性曲线、真菌结构饼图、丰度热点图及主坐标分析(PCoA)图[18],并找出不同肥瘦率金华猪的同肠段差异菌,与背膘厚做相关性分析,探究肠道真菌与脂肪沉积的关联性。
1.4 统计分析采用SPSS 22.0软件对数据进行统计分析,运用t检验(t-test)进行分析比较,P < 0.05为差异显著,结果以平均值表示。
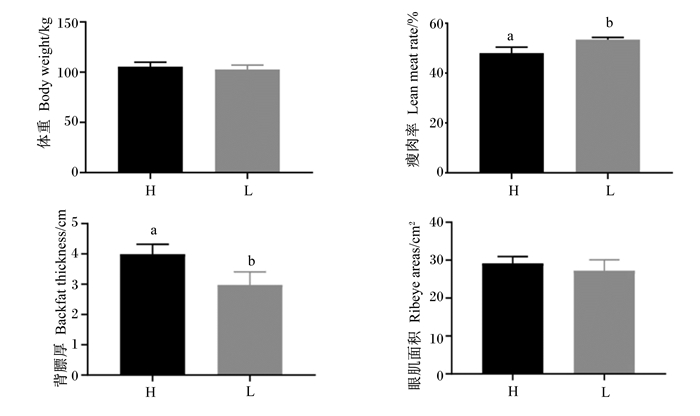
2 结果 2.1 不同肥瘦率金华猪的胴体品质由图 1可知,不同肥瘦率金华猪的瘦肉率与背膘厚存在显著差异(P < 0.05),H组金华猪瘦肉率比L组低5.49%,H组金华猪背膘厚比L组金华猪高25.36%;体重与眼肌面积H组与L组之间无显著差异(P>0.05)。
|
L: L组L group;H:H组H group。下图同the same as below。 图 1 不同肥瘦率金华猪的胴体品质 Fig. 1 Carcass quality of Jinhua pigs with different fatness rates |
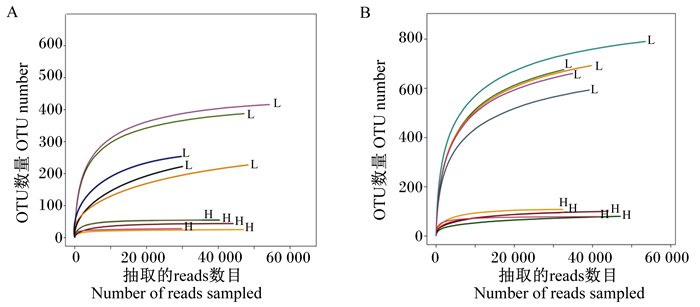
从稀释曲线(图 2)可以看出,H组和L组呈现出明显的差异。各样品覆盖度指数(Coverage index)为0.999~1.000,因此测序结果能代表样本的真实多样性组成。
|
图 2 不同肥瘦率金华猪回肠(A)和结肠(B)真菌稀释性曲线 Fig. 2 Fungi dilution curves in ileum (A) and colon (B) of Jinhua pigs with different fatness rates |
在OTU聚类97%相似水平下,根据香农指数、Chao1指数和辛普森指数对不同肥瘦率金华猪回肠、结肠真菌的丰富度和多样性进行分析。在回肠段(表 2),H组香浓指数、Chao1指数显著低于L组(P < 0.05)。在结肠段(表 3),H组OTU数量、香农指数、Chao1指数显著低于L组(P < 0.05)。
|
|
表 2 不同肥瘦率金华猪回肠真菌测序概况 Table 2 Fungi sequencing overview in ileum of Jinhua pigs with different fatness rates |
|
|
表 3 不同肥瘦率金华猪结肠真菌测序概况 Table 3 Fungi overview of sequencing in colon of Jinhua pigs with different fatness rates |
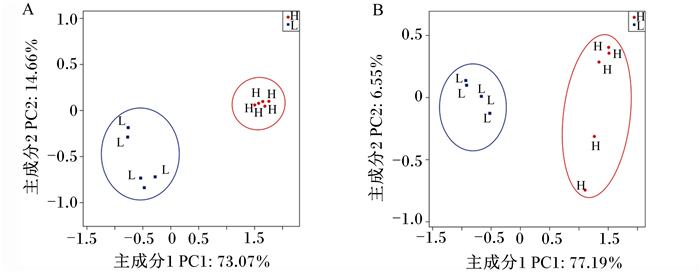
通过PCoA(图 3)发现,H组与L组呈现出明显不同的聚类,表明二者的菌群结构存在差异。
|
图 3 不同肥瘦率金华猪回肠(A)和结肠(B)真菌主坐标分析 Fig. 3 Fungi PCoA in ileum (A) and colon (B) of Jinhua pigs with different fatness rates |
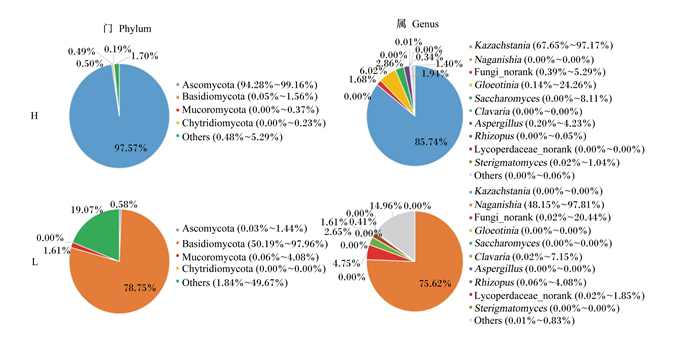
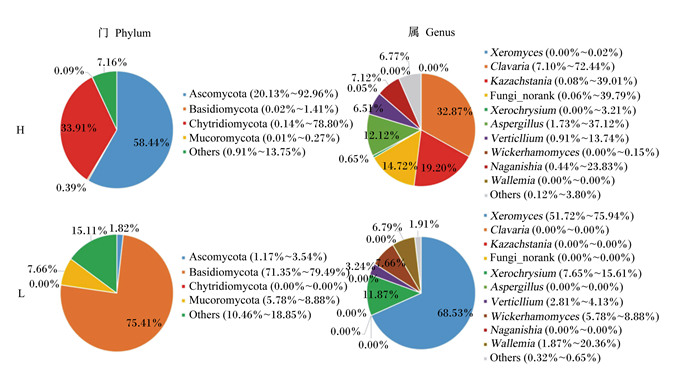
在门(phylum)水平上,回肠、结肠的优势真菌门均为子囊菌门(Ascomycota)、担子菌门(Basidiomycota)、毛霉菌门(Mucoromycota)和葫芦菌门(Chytridiomycota)。在回肠段(图 4),H组子囊菌门(Ascomycota)的相对丰度显著高于L组(97.57% vs 0.58%,P < 0.05),担子菌门(Basidiomycota)的相对丰度显著低于L组(0.49% vs 78.75%,P < 0.05)。在结肠段(图 5),H组子囊菌门(Ascomycota)的相对丰度显著高于L组(58.44% vs 1.82%,P < 0.05);H组担子菌门(Basidiomycota)的相对丰度显著低于L组(0.09% vs 75.41%,P < 0.05)。
|
Ascomycota:子囊菌门;Basidiomycota:担子菌门;Mucoromycota:毛霉菌门;Chytridiomycota:葫芦菌门;Kazachstania:扎斯坦酵母属;Naganishia:担子菌酵母属;Gloeotinia:内生真菌属;Saccharomyces:布拉酵母菌属;Clavaria:珊瑚菌属;Aspergillus:曲霉菌属;Rhizopus:根霉菌属;Sterigmatomyces:梗孢酵母属;Others:其他。 图 4 不同肥瘦率金华猪回肠真菌结构 Fig. 4 Fungi structure in ileum of Jinhua pigs with different fatness rates |
在属(genus)水平上,在回肠段(图 4),相对丰度最高的10个菌属为卡扎斯坦酵母属(Kazachstania)、担子菌酵母属(Naganishia)、Fungi_norank、内生真菌属(Gloeotinia)、布拉酵母菌属(Saccharomyces)、珊瑚菌属(Clavaria)、曲霉菌属(Aspergillus)、根霉菌属(Rhizopus)、Lycoperdaccae_norank、梗孢酵母属(Sterigmatomyces)。H组Kazachstania的相对丰度显著高于L组(85.74% vs 0.00%,P < 0.05),而Naganishia的相对丰度显著低于L组(0.00% vs 75.62%,P < 0.05)。在结肠段(图 5),相对丰度最高的10个菌属为耐干霉菌属(Xeromyces)、Clavaria、Kazachstania、Fungi_norank、金孢菌属(Xerochrysium)、Aspergillus、轮枝霉属(Verticllium)、威克汉姆酵母属(Wickerhamomyces)、Naganishia、节担菌属(Wallemia)。H组Clavaria的相对丰度显著高于L组(32.87% vs 0.00%,P < 0.05),而Xerochrysium的相对丰度显著低于L组(0.00% vs 68.53%,P < 0.05)。
|
Ascomycota子囊菌门;Basidiomycota:担子菌门;Chytridiomycota:葫芦菌门;Mucoromycota:毛霉菌门;Xeromyces:耐干霉菌属;Clavaria:珊瑚菌属;Kazachstania:扎斯坦酵母属;Xerochrysium:金孢菌属;Aspergillus:曲霉菌属;Verticllium:轮枝霉属;Wickerhamomyces:威克汉姆酵母属;Naganishia:担子菌酵母属;Wallemia:节担菌属;Others:其他。 图 5 不同肥瘦率金华猪结肠真菌结构 Fig. 5 Fungi structure in colon of Jinhua pigs with different fatness rates |
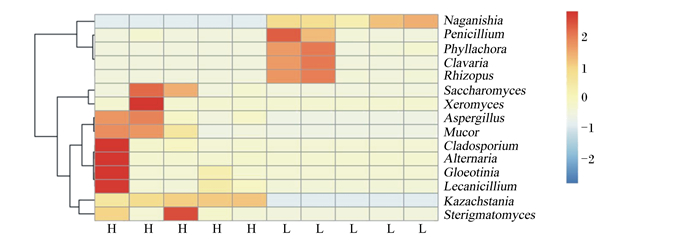
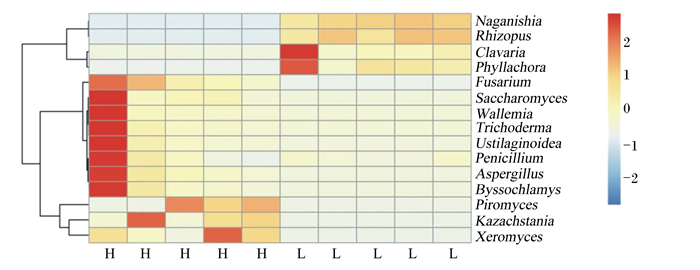
通过对相对丰度最高的15个优势真菌属进行热图(图 6和图 7)分析,发现2个肠段有2个共同的差异真菌属,Kazachstania在H组肠道中相对丰度较高,Naganishia在L组中相对丰度较高。
|
Naganishia:担子菌酵母属;Penicillium:青霉菌属;Phyllachora:黑痣菌属;Clavaria:珊瑚菌属;Rhizopus:根霉菌属;Saccharomyces:布拉酵母菌属;Xeromyces:耐干霉菌属;Aspergillus:曲霉菌属;Mucor:毛霉属;Cladosporium:枝孢属;Alternaria:链格孢属;Gloeotinia:内生真菌属;Lecanicillium:蜡蚧菌属;Kazachstania:扎斯坦酵母属;Sterigmatomyces:梗孢酵母属。 图 6 不同肥瘦率金华猪回肠真菌结构热图(属水平) Fig. 6 Heat map of fungi structure in ileum of Jinhua pigs with different fatness rates (at genus level) |
|
Naganishia:担子菌酵母属;Rhizopus:根霉菌属;Clavaria:珊瑚菌属;Phyllachora:黑痣菌属;Fusarium:镰刀菌属;Saccharomyces:布拉酵母菌属;Wallemia:节担菌属;Trichoderma:木霉菌属;Ustilaginoidea:绿核菌属;Penicillium:青霉菌属;Aspergillus:曲霉菌属;Byssochlamys:丝衣霉菌属;Piromyces:鞭毛菌属;Kazachstania:扎斯坦酵母属;Xeromyces:耐干霉菌属。 图 7 不同肥瘦率金华猪结肠真菌结构热图(属水平) Fig. 7 Heat map of fungi structure in colon of Jinhua pigs with different fatness rates (at genus level) |
对不同肥瘦率金华猪回肠、结肠优势菌真属与背膘厚进行相关性分析发现。Kazachstania、Saccharomyces、Aspergillus相对丰度与背膘厚均呈高度正相关(r=0.59~0.92);Naganishia、Clavaria、Rhizopus相对丰度均与背膘厚呈负相关(r=-0.88~-0.78)(表 4)。
|
|
表 4 不同肥瘦率金华猪相对丰度前10真菌属与背膘厚的相关性分析 Table 4 Correlation analysis between relative abundance of top 10 fungus genera and backfat thickness of Jinhua pigs with different fatness rates |
机体的肠道内生存着大量的细菌、真菌等生物,其数量庞大、种类众多,它们彼此依赖、相互制约,构成了相对稳定的微生态系统。越来越多的研究证实了肠道菌群在脂肪沉积中的作用。例如,Backhed等[19]通过将肥胖小鼠中的肠道菌群移植到无菌小鼠后发现即使减少食物供应,小鼠的体脂含量仍持续增加,可见肠道菌群对脂质代谢的作用。在最近的研究中,John等[20]指出,肠道微生物群落对肥胖的影响并不是简单的拟杆菌门与厚壁菌门的比例及其互作,真菌也可能影响代谢过程。在本试验中,我们证实不同肥瘦率金华猪的相同肠段真菌结构不一样,Alpha多样性分析结果显示,L组金华猪回肠、结肠真菌的香农指数、Chao1指数远高于H组,这与陈镜刚[21]探究金华猪与长白猪回肠、结肠菌群结构所得试验结果一致。肠道微生物中的大部分存在于结肠中,其数量为109~1012 CFU/mL[22-23],与此相一致,在本试验中结肠中真菌数量远高于回肠,且结肠中的优势真菌属差异大于回肠中。前人在小鼠中的研究中证明念珠菌是肠道真菌群落中最丰富的菌属之一[24],然而本试验中鉴定得出H组金华猪回肠优势真菌属为Kazachstania,L组金华猪回肠优势真菌属为Naganishia,H组金华猪结肠无优势真菌属,但L组金华猪结肠中优势真菌属为Xeromyces,其差异可能与猪的品种及其饲养条件有关。后续仍需要进一步的研究验证这3种优势真菌属是否与金华猪脂肪沉积机理有一定的关联性。
目前前人提出具有益生作用的真菌是布拉酵母菌(Saccharomyces boulardii)和汉逊德巴利酵母菌(Debaryomyces hanseni),其中,布拉酵母菌的临床试验显示出其具有调节肠道微生态的平衡的作用[25-26],并且可以改善腹泻等肠道炎症疾病[27-29]。Carbonetto等[30]和Hagman等[31]指出Kazachstania能在无氧和有氧条件下发酵己糖,并将己糖转化为乙酸和乙醇。在本试验中,发现Kazachstania相对丰度与背膘厚存在正相关关系(r=0.69~0.70),且该真菌属是H组金华猪回肠中的优势真菌属,这可能与该真菌属在肠道中分解大量的碳水化合物,提高了有机物质的利用率有关,最终导致宿主的脂肪沉积。Naganishia属于油质酵母,其产生的脂质富含单不饱和脂肪酸(monounsaturated fatty acids,MUFAs)和多不饱和脂肪酸(polyunsaturated fatty acid,PUFAs)[32-33],二者在细胞质中积累成疏水性脂质颗粒,可被细胞代谢用于膜生物合成原料和能量储备[34],MUFAs、PUFAs还参与机体免疫系统激活及机体脂肪代谢调控[35],这些都揭示了肠道真菌可通过其代谢产物参与机体的脂肪代谢调控。与此一致,本试验结果显示,Naganishia相对丰度与金华猪背膘厚存在负相关关系(r=-0.83~-0.79),表明有可能是Naganishia产生的MUFAs以及PUFAs等代谢物促进了机体免疫系统激活,并没有用来调控机体的脂肪代谢。综上可知,肠道真菌有可能参与了宿主脂质沉积等代谢活动,关于肠道真菌与脂肪代谢的具体机制,仍有待进一步探究。
4 结论① 金华猪回肠中优势真菌属为Kazachstania、Naganishia、Gloeotinia、Saccharomyces。
② 金华猪结肠中优势真菌属为Xeromyces、Clavaria、Kazachstania、Xerochrysium。
③ 金华猪肠道中Kazachstania相对丰度与背膘厚存在正相关关系(r=0.69~0.70),Naganishia相对丰度与背膘厚存在负相关关系(r=-0.83~-0.79),推测这2种真菌属可能与脂肪代谢存在一定的关系。
| [1] |
YANG M, SHI F H, LIU W, et al. Dapagliflozin modulates the fecal microbiota in a type 2 diabetic rat model[J]. Frontiers in Endocrinology, 2020, 11: 635. DOI:10.3389/fendo.2020.00635 |
| [2] |
HEINRITZ S N, WEISS E, EKLUND M, et al. Intestinal microbiota and microbial metabolites are changed in a pig model fed a high-fat/low-fiber or a low-fat/high-fiber diet[J]. PLoS One, 2016, 11(4): e0154329. DOI:10.1371/journal.pone.0154329 |
| [3] |
VALERIANO V D V, BALOLONG M P, KANG D K. Probiotic roles of Lactobacillus sp. in swine: insights from gut microbiota[J]. Journal of Applied Microbiology, 2017, 122(3): 554-567. DOI:10.1111/jam.13364 |
| [4] |
KIM H B, ISAACSON R E. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing[J]. Veterinary Microbiology, 2015, 177(3/4): 242-251. |
| [5] |
CANI P D. Human gut microbiome: hopes, threats and promises[J]. Gut, 2018, 67(9): 1716-1725. DOI:10.1136/gutjnl-2018-316723 |
| [6] |
OTT S J, KVHBACHER T, MUSFELDT M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity[J]. Scandinavian Journal of Gastroenterology, 2008, 43(7): 831-841. DOI:10.1080/00365520801935434 |
| [7] |
岳宏宇, 丛春莉, 李艳梅. 肠道微生态与肠道疾病关系的研究进展[J]. 中国真菌学杂志, 2020, 15(4): 240-243. YUE H Y, CONG C L, LI Y M. Research progress on the relationship between intestinal microecology and intestinal diseases[J]. Chinese Journal of Mycology, 2020, 15(4): 240-243 (in Chinese). DOI:10.3969/j.issn.1673-3827.2020.04.014 |
| [8] |
VÉTIZOU M, PITT J M, DAILLōRE R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota[J]. Science, 2015, 350(6264): 1079-1084. DOI:10.1126/science.aad1329 |
| [9] |
KÖHLER J R, HUBE B, PUCCIA R, et al. Fungi that infect humans[J]. Microbiology Spectrum, 2017, 5(3): FUNK-0014-2016. |
| [10] |
SHIMIZU Y, KONNO Y, TOMITA Y. Wickerhamomyces psychrolipolyticus f.a., sp. nov., a novel yeast species producing two kinds of lipases with activity at different temperatures[J]. International Journal of Systematic and Evolutionary Microbiology, 2020, 70(2): 1158-1165. DOI:10.1099/ijsem.0.003894 |
| [11] |
LEMOINNE S, KEMGANG A, BEN BELKACEM K, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis[J]. Gut, 2020, 69(1): 92-102. DOI:10.1136/gutjnl-2018-317791 |
| [12] |
RODRIGUES C S, CAMPOS C F, CUNHA C, et al. Metabolic regulation of innate immunity to fungal infection[M]//SILVESTRE R, TORRADO E. Metabolic interaction in infection. Cham: Springer, 2018: 403-420.
|
| [13] |
MIAO Z G, WANG L J, XU Z R, et al. Developmental changes of carcass composition, meat quality and organs in the Jinhua pig and Landrace[J]. Animal, 2009, 3(3): 468-473. DOI:10.1017/S1751731108003613 |
| [14] |
XIAO Y P, KONG F L, XIANG Y, et al. Comparative biogeography of the gut microbiome between Jinhua and Landrace pigs[J]. Scientific Reports, 2018, 8(1): 5985. DOI:10.1038/s41598-018-24289-z |
| [15] |
肖英平, 王军军, 李天天, 等. 金华猪和长白猪粪便微生物移植小鼠的肠道古菌结构分析[J]. 动物营养学报, 2017, 29(6): 1895-1903. XIAO Y P, WANG J J, LI T T, et al. Intestinal archaea community structure analysis of mice transplanted with Jinhua and Landrace pig feces[J]. Chinese Journal of Animal Nutrition, 2017, 29(6): 1895-1903 (in Chinese). DOI:10.3969/j.issn.1006-267x.2017.06.010 |
| [16] |
王远霞, 刘秀婷, 章啸君, 等. 金华猪回肠菌群结构和脂肪酸结合蛋白发育性变化与脂肪沉积的相关性研究[J]. 畜牧兽医学报, 2021, 52(3): 723-732. WANG Y X, LIU X T, ZHANG X J, et al. The developmental changes of ileal microbiota and fatty acid binding proteins and its correlation with fat deposition in Jinhua pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(3): 723-732 (in Chinese). |
| [17] |
ORGIAZZI A, LUMINI E, NILSSON R H, et al. Unravelling soil fungal communities from different Mediterranean land-use backgrounds[J]. PLoS One, 2012, 7(4): e34847. DOI:10.1371/journal.pone.0034847 |
| [18] |
JIANG X T, PENG X, DENG G H, et al. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland[J]. Microbial Ecology, 2013, 66(1): 96-104. DOI:10.1007/s00248-013-0238-8 |
| [19] |
BÄCKHED F, DING H, WANG T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(44): 15718-15723. DOI:10.1073/pnas.0407076101 |
| [20] |
JOHN G K, MULLIN G E. The gut microbiome and obesity[J]. Current Oncology Reports, 2016, 18(7): 45. DOI:10.1007/s11912-016-0528-7 |
| [21] |
陈镜刚. 金华猪与长白猪肠道消化酶活性、菌群区系与短链脂肪酸含量差异比较[D]. 硕士学位论文. 南京: 南京农业大学, 2016. CHEN J G. Comparison on intestinal digestive enzyme actinity, microflora and short chain fatty acid between Jinhua pigs and Landrace[D]. Master's Thesis. Nanjing: Nanjing Agricultural University, 2016. (in Chinese) |
| [22] |
徐娥, 杨华, 刘秀婷, 等. 大约克猪肠道不同部位的菌群结构和短链脂肪酸含量研究[J]. 动物营养学报, 2019, 31(10): 4509-4518. XU E, YANG H, LIU X T, et al. Study on bacterial community structure and short chain fatty acid content in different parts of intestines of Yorkshire pigs[J]. Chinese Journal of Animal Nutrition, 2019, 31(10): 4509-4518 (in Chinese). |
| [23] |
BLAUT M, CLAVEL T. Metabolic diversity of the intestinal microbiota: implications for health and disease[J]. The Journal of Nutrition, 2007, 137(3): 751S-755S. DOI:10.1093/jn/137.3.751S |
| [24] |
DOLLIVE S, CHEN Y Y, GRUNBERG S, et al. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment[J]. PLoS One, 2013, 8(8): e71806. DOI:10.1371/journal.pone.0071806 |
| [25] |
唐圆, 贺璐, 谢果珍, 等. 肠道真菌研究进展[J]. 中国微生态学杂志, 2019, 31(11): 1341-1346. TANG Y, HE L, XIE G Z, et al. Progress in research on intestinal fungi[J]. Chinese Journal of Microecology, 2019, 31(11): 1341-1346 (in Chinese). |
| [26] |
郭抗萧, 谭周进, 谢梦洲, 等. 超微七味白术散与酵母菌协同治疗小鼠菌群失调腹泻[J]. 应用与环境生物学报, 2015, 21(1): 61-67. GUO K X, TAN Z J, XIE M Z, et al. The synergic effect of ultra-micro powder Qiweibaizhusan combined with yeast on dysbacteriotic diarrhea mice[J]. Chinese Journal of Applied & Environmental Biology, 2015, 21(1): 61-67 (in Chinese). |
| [27] |
MCFARLAND L V. Meta-analysis of probiotics for the prevention of traveler's diarrhea[J]. Travel Medicine and Infectious Disease, 2007, 5(2): 97-105. DOI:10.1016/j.tmaid.2005.10.003 |
| [28] |
VANDENPLAS Y, DE GREEF E, DEVREKER T, et al. Probiotics and prebiotics in infants and children[J]. Current Infectious Disease Reports, 2013, 15(3): 251-262. DOI:10.1007/s11908-013-0334-4 |
| [29] |
TERCIOLO C, DOBRIC A, OUAISSI M, et al. Saccharomyces boulardii CNCM I-745 restores intestinal barrier integrity by regulation of E-cadherin recycling[J]. Journal of Crohn's & Colitis, 2017, 11(8): 999-1010. |
| [30] |
CARBONETTO B, NIDELET T, GUEZENEC S, et al. Interactions between Kazachstania humilis yeast species and lactic acid bacteria in sourdough[J]. Microorganisms, 2020, 8(2): 240. DOI:10.3390/microorganisms8020240 |
| [31] |
HAGMAN A, SÄLL T, COMPAGNO C, et al. Yeast "make-accumulate-consume" life strategy evolved as a multi-step process that predates the whole genome duplication[J]. PLoS One, 2013, 8(7): e68734. DOI:10.1371/journal.pone.0068734 |
| [32] |
FILIPPUCCI S, TASSELLI G, SCARDUA A, et al. Study of Holtermanniella wattica, Leucosporidium creatinivorum, Naganishia adeliensis, Solicoccozyma aeria, and Solicoccozyma terricola for their lipogenic aptitude from different carbon sources[J]. Biotechnology for Biofuels, 2016, 9: 259. DOI:10.1186/s13068-016-0672-1 |
| [33] |
PAPANIKOLAOU S, CHEVALOT I, KOMAITIS M, et al. Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures[J]. Applied Microbiology and Biotechnology, 2002, 58(3): 308-312. DOI:10.1007/s00253-001-0897-0 |
| [34] |
PAPANIKOLAOU S, CHEVALOT I, KOMAITIS M, et al. Kinetic profile of the cellular lipid composition in an oleaginous Yarrowia lipolytica capable of producing a cocoa-butter substitute from industrial fats[J]. Antonie van Leeuwenhoek, 2001, 80(3/4): 215-224. DOI:10.1023/A:1013083211405 |
| [35] |
CÂNDIDO F G, VALENTE F X, GRZEŚKOWIAK Ł M, et al. Impact of dietary fat on gut microbiota and low-grade systemic inflammation: mechanisms and clinical implications on obesity[J]. International Journal of Food Sciences and Nutrition, 2018, 69(2): 125-143. DOI:10.1080/09637486.2017.1343286 |




